AI Tool SCORPIO Predicts Cancer Immunotherapy Response Using Routine Blood Tests
4 Sources
4 Sources
[1]
New AI tool uses routine blood tests to predict immunotherapy response for many cancers
Doctors around the world may soon have access to a new tool that could better predict whether individual cancer patients will benefit from immune checkpoint inhibitors -- a type of immunotherapy -- using only routine blood tests and clinical data. The artificial intelligence-based model, dubbed SCORPIO, was developed by a team of researchers from Memorial Sloan Kettering Cancer Center (MSK) and the Tisch Cancer Institute at Mount Sinai. The model is not only cheaper and more accessible, it's significantly better at predicting outcomes than the two current biomarkers approved by the U.S. Food and Drug Administration (FDA), according to findings published in Nature Medicine. "Immune checkpoint inhibitors are a very powerful tool against cancer, but they don't yet work for most patients," says study co-senior author Luc Morris, MD, a surgeon and research lab director at MSK. "These drugs are expensive, and they can come with serious side effects." So the key is patient selection -- matching the drugs with patients who are most likely to benefit, Dr. Morris says. "There are some existing tools that predict whether tumors will respond to these drugs, but they tend to rely on advanced genomic testing that is not widely available around the world," he adds. "We wanted to develop a model that can help guide treatment decisions using widely available data, such as routine blood tests." Collaborating to make checkpoint inhibitor therapy work for more cancer patients Checkpoint inhibitors target the immune system rather than the cancer itself. These drugs work by taking the brakes off immune cells, allowing them to better fight cancer. MSK clinicians and scientists played a key role in bringing the new class of drugs to patients. The new study was jointly overseen by Dr. Morris and Diego Chowell, Ph.D., an Assistant Professor of Immunology and Immunotherapy, Oncological Sciences, and Artificial Intelligence and Human Health at the Icahn School of Medicine at Mount Sinai, and a former postdoctoral fellow at MSK. Q&A with Dr. Morris Memorial Sloan Kettering Cancer Center spoke with Dr. Morris about the team's prediction model and next steps for the research. Why did you develop this new model to predict checkpoint inhibitor response? It was clear there was room for improvement. There are currently two FDA-approved biomarkers for predicting response to checkpoint inhibitors: tumor mutational burden (the number of mutations in a tumor) and PD-L1 immunohistochemistry (evaluating the expression of the programmed death-ligand 1 protein in tumor samples). Both require samples of the tumor to be collected. Meanwhile, genomic testing to assess mutations is expensive and not available everywhere, and there is a lot of variability evaluating PD-L1 expression. Instead, our model relies on readily available clinical data, including routine blood tests performed in clinics around the world -- the complete blood count and the comprehensive metabolic profile. We found that our model outperforms the currently used tests in the clinic. The simplicity and affordability of this new approach could help ensure more equitable access to care while also reducing costs and helping ensure patients receive treatments most likely to benefit them individually -- whether that ends up being a checkpoint inhibitor or some other type of therapy. How was the model developed? SCORPIO was initially developed by our team by collecting data from MSK patients, because of the length and depth of experience oncologists here have treating patients with these drugs. Collaborating with the team at Mount Sinai, we used a type of artificial intelligence called ensemble machine learning, which combines several tools to look for patterns in clinical data from blood tests and treatment outcomes. The model was developed using a rich resource of retrospective data from more than 2,000 patients from MSK who had been treated with checkpoint inhibitors, representing 17 different types of cancer. The model was then tested using data from 2,100 additional MSK patients to verify that it was able to predict outcomes with high accuracy. Next, we applied the model to nearly 4,500 patients treated with checkpoint inhibitors in 10 different phase 3 clinical trials from around the world. Further validation was done with additional data from nearly 1,200 patients treated at Mount Sinai. In total, the study included nearly 10,000 patients across 21 different cancer types -- representing the largest dataset in cancer immunotherapy to date. We did this extensive testing and validation because our goal was not just to develop a predictive model, but to develop one that would be widely applicable to patients and physicians in different locations. What are the next steps? We plan to collaborate with hospitals and cancer centers around the world to test the model with additional data from a wider variety of clinical settings. The feedback we receive will help us to continue to optimize the model. Additionally, work is underway to develop an interface that is readily accessible by clinicians, regardless of where they're located.
[2]
New AI Tool Uses Routine Blood Tests to Predict Im | Newswise
Doctors around the world may soon have access to a new tool that could better predict whether individual cancer patients will benefit from immune checkpoint inhibitors -- a type of immunotherapy -- using only routine blood tests and clinical data. The artificial intelligence-based model, dubbed SCORPIO, was developed by a team of researchers from Memorial Sloan Kettering Cancer Center (MSK) and the Tisch Cancer Institute at Mount Sinai. The model is not only cheaper and more accessible, it's significantly better at predicting outcomes than the two current biomarkers approved by the U.S. Food and Drug Administration (FDA), according to findings published January 6 in Nature Medicine. "Immune checkpoint inhibitors are a very powerful tool against cancer, but they don't yet work for most patients," says study co-senior author Luc Morris, MD, a surgeon and research lab director at MSK. "These drugs are expensive, and they can come with serious side effects." So the key is patient selection -- matching the drugs with patients who are most likely to benefit, Dr. Morris says. "There are some existing tools that predict whether tumors will respond to these drugs, but they tend to rely on advanced genomic testing that is not widely available around the world," he adds. "We wanted to develop a model that can help guide treatment decisions using widely available data, such as routine blood tests." Checkpoint inhibitors target the immune system rather than the cancer itself. These drugs work by taking the brakes off immune cells, allowing them to better fight cancer. MSK clinicians and scientists played a key role in bringing the new class of drugs to patients. The new study was jointly overseen by Dr. Morris and Diego Chowell, PhD, an Assistant Professor of Immunology and Immunotherapy, Oncological Sciences, and Artificial Intelligence and Human Health at the Icahn School of Medicine at Mount Sinai, and a former postdoctoral fellow at MSK. We spoke with Dr. Morris about the team's prediction model and next steps for the research: It was clear there was room for improvement. There are currently two FDA-approved biomarkers for predicting response to checkpoint inhibitors: tumor mutational burden (the number of mutations in a tumor) and PD-L1 immunohistochemistry (evaluating the expression of the programmed death-ligand 1 protein in tumor samples). Both require samples of the tumor to be collected. Meanwhile, genomic testing to assess mutations is expensive and not available everywhere, and there is a lot of variability evaluating PD-L1 expression. Instead, our model relies on readily available clinical data, including routine blood tests performed in clinics around the world -- the complete blood count and the comprehensive metabolic profile. And we found that our model outperforms the currently used tests in the clinic. The simplicity and affordability of this new approach could help ensure more equitable access to care while also reducing costs and helping ensure patients receive treatments most likely to benefit them individually -- whether that ends up being a checkpoint inhibitor or some other type of therapy. SCORPIO was initially developed by our team, by collecting data from MSK patients, because of the length and depth of experience oncologists here have treating patients with these drugs. Collaborating with the team at Mount Sinai, we used a type of artificial intelligence called 'ensemble machine learning,' which combines several tools to look for patterns in clinical data from blood tests and treatment outcomes. The model was developed using a rich resource of retrospective data from more than 2,000 patients from MSK who had been treated with checkpoint inhibitors, representing 17 different types of cancer. The model was then tested using data from 2,100 additional MSK patients to verify that it was able to predict outcomes with high accuracy. Next, we applied the model to nearly 4,500 patients treated with checkpoint inhibitors in 10 different phase 3 clinical trials from around the world. Further validation was done with additional data from nearly 1,200 patients treated at Mount Sinai. In total, the study includes nearly 10,000 patients across 21 different cancer types -- representing the largest dataset in cancer immunotherapy to date. We did this extensive testing and validation because our goal was not just to develop a predictive model, but to develop one that would be widely applicable to patients and physicians in different locations. We plan to collaborate with hospitals and cancer centers around the world to test the model with additional data from a wider variety of clinical settings. The feedback we receive will help us to continue to optimize the model. Additionally, work is underway to develop an interface that is readily accessible by clinicians, regardless of where they're located. The study represents the work of 37 authors. The research was led by co-first authors Seong-Keun Yoo, PhD, and Byuri Angela Cho, PhD, research fellows in the Chowell Lab; Conall Fitzgerald , MD, a research fellow in the Morris Lab who is now a consultant head and neck surgeon at St James's Hospital and Trinity College Dublin, Ireland; and Bailey G. Fitzgerald, MD, a medical oncology fellow at Mount Sinai. Please refer to the study for the full list. This research was supported in part by the National Institutes of Health (R01 DE027738, R01 CA283469, U01CA282114, P30 CA008748, P30 CA196521), the Department of Defense, the Geoffrey Beene Cancer Research Center at MSK, Cycle for Survival, the Jayme and Peter Flowers Fund, the Sebastian Nativo Fund, and the Alexander and Alexandrine Sinsheimer Foundation. Please refer to the study for a full list of funders. Several of the authors have filed a provisional patent application for using routine blood tests and clinical variables to predict cancer immunotherapy response. Several are also co-inventors on a patent by MSK for using tumor mutational burden to predict immunotherapy response, licensed to Personal Genome Diagnostics. Another group are co-inventors on a patent filed jointly by Cleveland Clinic and MSK for a multimodal machine learning model to predict immunotherapy response, licensed to Tempus. A number of the authors also report consulting work for pharmaceutical companies, unrelated to the current research. Please refer to the study for a full list of disclosures.
[3]
AI-based SCORPIO tool offers better cancer immunotherapy predictions
Memorial Sloan Kettering Cancer CenterJan 6 2025 Doctors around the world may soon have access to a new tool that could better predict whether individual cancer patients will benefit from immune checkpoint inhibitors - a type of immunotherapy - using only routine blood tests and clinical data. The artificial intelligence-based model, dubbed SCORPIO, was developed by a team of researchers from Memorial Sloan Kettering Cancer Center (MSK) and the Tisch Cancer Institute at Mount Sinai. The model is not only cheaper and more accessible, it's significantly better at predicting outcomes than the two current biomarkers approved by the U.S. Food and Drug Administration (FDA), according to findings published January 6 in Nature Medicine. Immune checkpoint inhibitors are a very powerful tool against cancer, but they don't yet work for most patients. These drugs are expensive, and they can come with serious side effects." Luc Morris, MD, study co-senior author, surgeon and research lab director at MSK So the key is patient selection - matching the drugs with patients who are most likely to benefit, Dr. Morris says. "There are some existing tools that predict whether tumors will respond to these drugs, but they tend to rely on advanced genomic testing that is not widely available around the world," he adds. "We wanted to develop a model that can help guide treatment decisions using widely available data, such as routine blood tests." Collaborating to make checkpoint inhibitor therapy work for more cancer patients Checkpoint inhibitors target the immune system rather than the cancer itself. These drugs work by taking the brakes off immune cells, allowing them to better fight cancer. MSK clinicians and scientists played a key role in bringing the new class of drugs to patients. The new study was jointly overseen by Dr. Morris and Diego Chowell, PhD, an Assistant Professor of Immunology and Immunotherapy, Oncological Sciences, and Artificial Intelligence and Human Health at the Icahn School of Medicine at Mount Sinai, and a former postdoctoral fellow at MSK. Q&A with Dr. Morris We spoke with Dr. Morris about the team's prediction model and next steps for the research: Why did you develop this new model to predict checkpoint inhibitor response? It was clear there was room for improvement. There are currently two FDA-approved biomarkers for predicting response to checkpoint inhibitors: tumor mutational burden (the number of mutations in a tumor) and PD-L1 immunohistochemistry (evaluating the expression of the programmed death-ligand 1 protein in tumor samples). Both require samples of the tumor to be collected. Meanwhile, genomic testing to assess mutations is expensive and not available everywhere, and there is a lot of variability evaluating PD-L1 expression. Instead, our model relies on readily available clinical data, including routine blood tests performed in clinics around the world - the complete blood count and the comprehensive metabolic profile. And we found that our model outperforms the currently used tests in the clinic. The simplicity and affordability of this new approach could help ensure more equitable access to care while also reducing costs and helping ensure patients receive treatments most likely to benefit them individually - whether that ends up being a checkpoint inhibitor or some other type of therapy. How was the model developed? SCORPIO was initially developed by our team, by collecting data from MSK patients, because of the length and depth of experience oncologists here have treating patients with these drugs. Collaborating with the team at Mount Sinai, we used a type of artificial intelligence called 'ensemble machine learning,' which combines several tools to look for patterns in clinical data from blood tests and treatment outcomes. The model was developed using a rich resource of retrospective data from more than 2,000 patients from MSK who had been treated with checkpoint inhibitors, representing 17 different types of cancer. The model was then tested using data from 2,100 additional MSK patients to verify that it was able to predict outcomes with high accuracy. Next, we applied the model to nearly 4,500 patients treated with checkpoint inhibitors in 10 different phase 3 clinical trials from around the world. Further validation was done with additional data from nearly 1,200 patients treated at Mount Sinai. In total, the study includes nearly 10,000 patients across 21 different cancer types - representing the largest dataset in cancer immunotherapy to date. We did this extensive testing and validation because our goal was not just to develop a predictive model, but to develop one that would be widely applicable to patients and physicians in different locations. What are the next steps? We plan to collaborate with hospitals and cancer centers around the world to test the model with additional data from a wider variety of clinical settings. The feedback we receive will help us to continue to optimize the model. Additionally, work is underway to develop an interface that is readily accessible by clinicians, regardless of where they're located. Additional authors, funding, and disclosures The study represents the work of 37 authors. The research was led by co-first authors Seong-Keun Yoo, PhD, and Byuri Angela Cho, PhD, research fellows in the Chowell Lab; Conall Fitzgerald , MD, a research fellow in the Morris Lab who is now a consultant head and neck surgeon at St James's Hospital and Trinity College Dublin, Ireland; and Bailey G. Fitzgerald, MD, a medical oncology fellow at Mount Sinai. Please refer to the study for the full list. This research was supported in part by the National Institutes of Health (R01 DE027738, R01 CA283469, U01CA282114, P30 CA008748, P30 CA196521), the Department of Defense, the Geoffrey Beene Cancer Research Center at MSK, Cycle for Survival, the Jayme and Peter Flowers Fund, the Sebastian Nativo Fund, and the Alexander and Alexandrine Sinsheimer Foundation. Please refer to the study for a full list of funders. Several of the authors have filed a provisional patent application for using routine blood tests and clinical variables to predict cancer immunotherapy response. Several are also co-inventors on a patent by MSK for using tumor mutational burden to predict immunotherapy response, licensed to Personal Genome Diagnostics. Another group are co-inventors on a patent filed jointly by Cleveland Clinic and MSK for a multimodal machine learning model to predict immunotherapy response, licensed to Tempus. A number of the authors also report consulting work for pharmaceutical companies, unrelated to the current research. Memorial Sloan Kettering Cancer Center Journal reference: Yoo, S.-K., et al. (2025). Prediction of checkpoint inhibitor immunotherapy efficacy for cancer using routine blood tests and clinical data. Nature Medicine. doi.org/10.1038/s41591-024-03398-5.
[4]
Prediction of checkpoint inhibitor immunotherapy efficacy for cancer using routine blood tests and clinical data - Nature Medicine
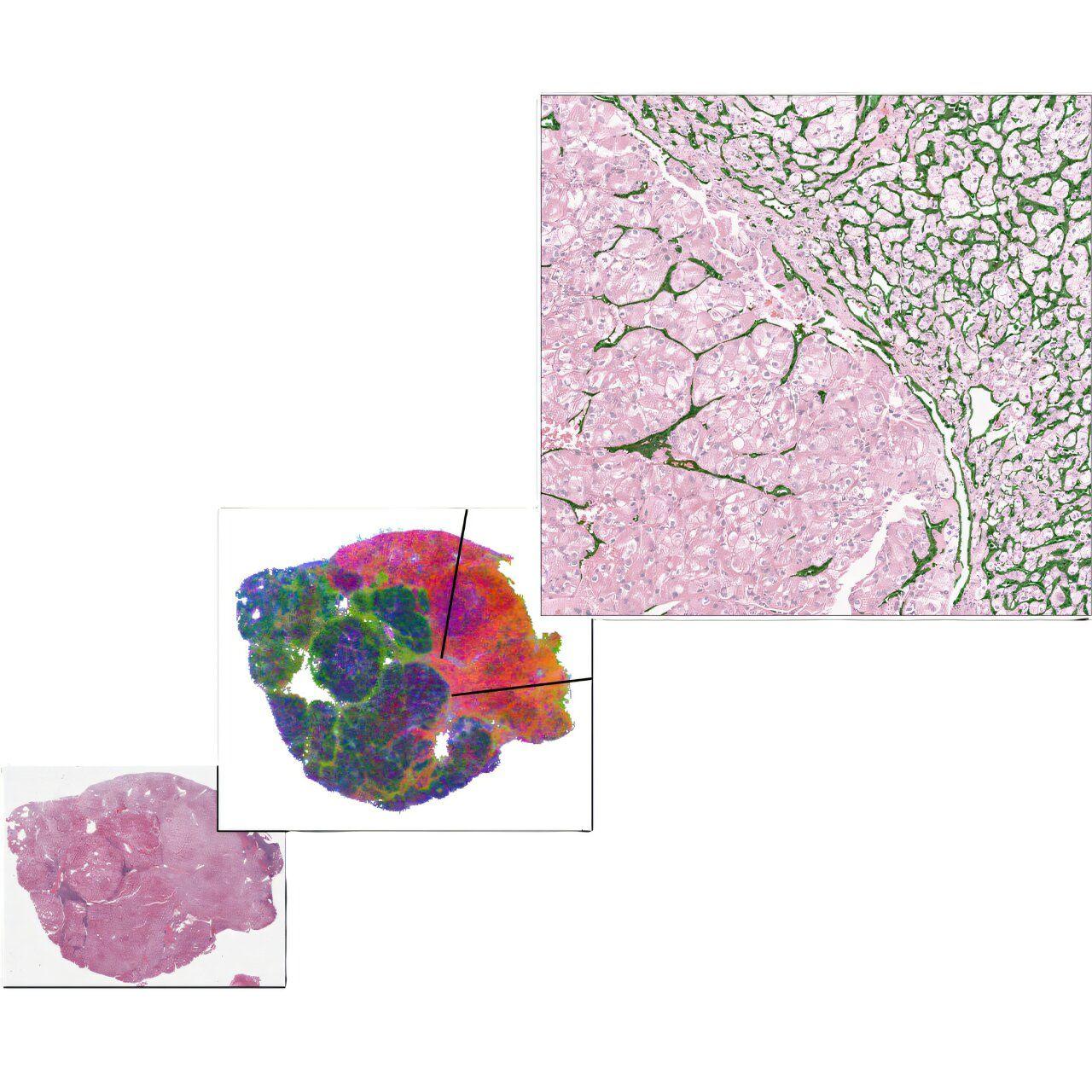
Machine learning, a branch of artificial intelligence, enables algorithms to learn from data, identify key patterns and make predictions14. These models have already shown success in various biomedical fields7,9,15,16,17,18,19. In this study, we explored whether a machine learning system could predict ICI outcomes using routine blood tests and standard clinical variables. We trained, tested and externally tested a machine learning model on three real-world cohorts and 10 global phase 3 clinical trial cohorts to predict ICI efficacy. This study included 9,745 patients across 21 cancer types treated with ICIs from Memorial Sloan Kettering Cancer Center (MSKCC), Mount Sinai Health System (MSHS) and 10 global phase 3 clinical trials (Fig. 1a, Extended Data Tables 1-3 and Supplementary Fig. 1). To develop the model, we first retrospectively collected data from 2,035 patients across 17 cancer types treated with ICIs between 2014 and 2019 at MSKCC (hereafter referred to as MSK-I), which were randomly divided into a training set (n = 1,628) and hold-out test set (n = 407) with 80:20 ratio. We developed machine learning models using the training set from this cohort and then tested the models in the hold-out test sets. We then further tested the model on an independent cohort of additional 2,104 ICI-treated patients from MSKCC (hereafter referred to as MSK-II). The MSK-II cohort was collected after initial model development, and identical inclusion and exclusion criteria were used as the MSK-I cohort, but the years of eligibility were expanded to patients treated between 2011 and 2020. We then externally tested the model on 4,447 ICI-treated patients in 10 global phase 3 clinical trials. Further external testing of the model was performed on a real-world cohort of 1,159 patients treated with ICIs between 2011 and 2019 at MSHS, a large comprehensive health system serving a diverse patient population across the New York metropolitan region. We also analyzed 6,629 patients treated for cancer at MSKCC who did not receive ICI (hereafter referred to as MSK non-ICI; Extended Data Table 4). For details on patient inclusion and exclusion criteria, see Methods and Supplementary Figs. 2 and 3. Patients were treated with inhibitors of PD-1 (n = 3,793), PD-L1 (n = 5,253), CTLA-4 (n = 72) or combinations of more than one drug (n = 627), including anti-CTLA-4 with anti-PD-1, anti-CTLA-4 with anti-PD-L1, anti-CTLA-4 with anti-PD-1 and anti-PD-L1 and anti-PD-1 and anti-PD-L1. The median follow-up duration for each cohort was: 25.38 months (interquartile range (IQR) 13.50-45.01) for the training set, 27.37 months (IQR 13.68-49.58) for the hold-out test set, 9.42 months (IQR 3.10-20.67) for the MSK-II cohort, 8.84 months (IQR 2.75-28.47) for the MSHS cohort and 13.64 months (IQR 6.72-19.86) across the clinical trials. The 10 clinical trial cohorts included patients from 12 experimental arms treated with atezolizumab (anti-PD-L1): IMbrave150 (ref. ), IMspire150 (ref. ), IMmotion151 (ref. ), IMvigor211 (ref. ), IMpower133 (ref. ), IMpower130 (ref. ), IMpower131 (atezolizumab plus carboplatin and nanoparticle ALB-bound paclitaxel (ACNP)), IMpower131 (atezolizumab plus carboplatin and paclitaxel (ACP)), IMpower132 (ref. ), IMpower150 (atezolizumab plus bevacizumab, carboplatin and paclitaxel (ABCP)), IMpower150 (ACP) and OAK. The median follow-up duration of each clinical trial cohort is provided in Extended Data Table 3. We analyzed bladder cancer, hepatobiliary cancer, melanoma, non-small cell lung cancer (NSCLC), renal cell carcinoma (RCC) and small cell lung cancer (SCLC) as separate cancer types as they were collected in all available cohorts. The remaining cancer types were grouped as 'Others' in each cohort. We retrospectively collected clinical variables and standardized measurements from routine laboratory blood tests performed on the date of, or no more than 30 days before, the first ICI infusion (Fig. 1b and Supplementary Table 1). In the MSKCC cohorts, TMB was collected from patients' tumors based on the FDA-authorized MSK-IMPACT platform. In the clinical trial cohorts, PD-L1 immunostaining data using the SP142 or SP263 clones (Ventana Medical Systems) were collected (Methods). The two primary outcomes were overall survival and a treatment effect, measured as clinical benefit. Overall survival was measured from the first ICI infusion to death from any cause, with the first line used for patients who received multiple ICI treatments. For clinical trial cohorts, overall survival was measured from randomization to death from any cause. Patients alive at the time of review were censored at their last contact. Clinical benefit was defined as a patient's tumor showing a complete response (CR), partial response (PR) or stable disease (SD) without progression for at least 6 months after the first infusion of ICI, as in prior studies. Patients whose tumors showed PD or SD for <6 months after the first ICI infusion were classified as having no clinical benefit. CR, PR, SD and PD were based on RECIST v1.1 criteria. Both primary outcomes were available in the MSKCC and clinical trial cohorts, but only overall survival data were available in the MSHS cohort. For a description of clinical features and outcomes, see Methods. Before model training, we performed feature selection analyses on the training set to identify features associated with the target outcomes of ICI treatment (Fig. 1b and Supplementary Fig. 4). We developed two machine learning models using demographic, clinical and routine laboratory blood test data to predict outcomes after ICI administration, with one trained to predict overall survival and the other trained to predict clinical benefit (CR, PR and SD ≥6 months), and selected the one that performed the best in the hold-out test set (Fig. 1c). Each model consisted of an ensemble of three algorithms with soft-voting. A five-fold cross-validation (CV) was used to optimize each algorithm's hyperparameters during training. During model training, the training set was divided into five equal-sized folds, each containing the same proportion of data. The algorithm then underwent five iterations of training and evaluation. In each iteration, four folds were used for training, and one fold was used for validation. Model performance was assessed using the concordance index (C-index) for overall survival and the area under the receiver operating characteristic curve (AUC) for clinical benefit. The performance metrics from the five iterations were averaged to obtain a single performance measurement. This process was repeated for all possible hyperparameter combinations, and the hyperparameter with the highest performance metric was selected as the optimal hyperparameter. The model trained to predict overall survival, SCORPIO (Standard Clinical and labOratory featuRes for Prognostication of Immunotherapy Outcomes), calculates a risk score ranging from 0 to 1, where a higher score indicates a higher probability of a poor outcome (that is, lack of efficacy or early death) after ICI administration. This model was trained using 33 features significantly associated with overall survival, identified through feature selection analysis (Supplementary Fig. 4a and Supplementary Table 2). Similarly, SCORPIO-CB, trained to predict clinical benefit, generates a probability score from 0 to 1, with a higher score indicating a higher likelihood of clinical benefit. This model was trained with 22 features significantly associated with clinical benefit, as identified in the feature selection analysis (Supplementary Fig. 4b and Supplementary Table 2). The performance of the two models was assessed using time-dependent AUC (AUC(t)) for overall survival and AUC for clinical benefit. For details of the machine learning system, see Methods. For the primary analysis of prognosticating clinical outcomes, patients were stratified into high-risk, moderate-risk and low-risk groups based on the first and third quartiles of the risk scores observed in the training set (Supplementary Fig. 5). The Cox proportional hazards regression tested the association of risk scores with overall survival and the Fisher's exact test compared clinical benefit rates across the three risk groups. For details of the statistical analysis, see Methods. In the hold-out test data, SCORPIO, the machine learning model trained to predict overall survival, prognosticated overall survival at 6, 12, 18, 24 and 30 months following ICI with a median pan-cancer AUC(t) of 0.763 (Fig. 2). SCORPIO outperformed SCORPIO-CB and TMB in predicting overall survival, as shown by AUC(t) values (Supplementary Fig. 6a). It also predicted clinical benefit with a pan-cancer AUC of 0.714, surpassing SCORPIO-CB (pan-cancer AUC 0.701) and TMB (pan-cancer AUC 0.546; Fig. 2 and Supplementary Fig. 6b). SCORPIO consistently outperformed both SCORPIO-CB and TMB across all cancer types (Supplementary Figs. 6 and 7). To determine whether cancer-type-specific models provide better predictive value than SCORPIO, a pan-cancer model, we developed models trained on data specific to each cancer type. First, we conducted feature selection analyses and model training separately for each cancer type. Among the 17 cancer types in the training set, we identified 10 with features significantly associated with overall survival (Supplementary Table 2). We then trained 10 models and compared their performance to SCORPIO in the hold-out test set. SCORPIO outperformed most of the cancer-type-specific models in predicting both overall survival and clinical benefit (Supplementary Fig. 8). This indicates that SCORPIO trained on the large pan-cancer data successfully learned relevant relationships across cancer types. Next, we compared SCORPIO's performance to nine machine learning models from Vanguri et al., which predict ICI efficacy in patients with NSCLC using uni-, bi- or multi-modal data (radiology, pathology, tumor genetics and PD-L1 scoring). SCORPIO outperformed these models in prognosticating overall survival (Supplementary Fig. 9a) and showed comparable performance in predicting clinical benefit, even though it was trained on simpler, more accessible pan-cancer data (Supplementary Fig. 9b). In the hold-out test data, the three risk groups (low-risk, moderate-risk and high-risk) showed significantly different overall survival (Supplementary Fig. 10a). Across tumor types, the hazard ratios (HRs) for death compared to the high-risk group were 0.25 (95% confidence interval (CI), 0.18-0.34) for the low-risk group and 0.48 (95% CI, 0.37-0.63) for the moderate-risk group. Furthermore, the clinical benefit rates significantly differed in each risk group across tumor types - low-risk, 55.96%; moderate-risk, 28.64%; high-risk, 12.12% (P = 3.22 × 10; Supplementary Fig. 10b). We then sought to test SCORPIO on the independent real-world MSK-II cohort. In this cohort, SCORPIO prognosticated overall survival at 6, 12, 18, 24 and 30 months following ICI with a median pan-cancer AUC(t) of 0.759 (Fig. 2). It also predicted clinical benefit from ICI with a pan-cancer AUC of 0.641. In accordance with the results from the hold-out test data, SCORPIO outperformed TMB based on both AUC(t) and AUC (Supplementary Figs. 11 and 12). The three risk groups had significantly different overall survival (Fig. 3a and Supplementary Fig. 13a). Across tumor types, HRs for death in the low-risk and moderate-risk groups compared to the high-risk group were 0.16 (95% CI, 0.14-0.19) and 0.38 (95% CI, 0.34-0.43), respectively. Furthermore, the clinical benefit rates significantly differed in each risk group across tumor types -- low-risk, 65.09%; moderate-risk, 52.20%; high-risk, 32.89% (P = 2.35 × 10; Fig. 3b and Supplementary Fig. 13b). In both internal test datasets, the association between risk groups and clinical outcomes was independent of the line of therapy in which ICI was administered, sex, age, Eastern Cooperative Oncology Group performance status (ECOG-PS), microsatellite instability (MSI) status and TMB (Supplementary Figs. 14 and 15). To determine whether SCORPIO is specifically prognostic for ICI efficacy or generally prognostic for patients with cancer regardless of treatment, we analyzed a cohort of non-ICI-treated patients from MSKCC. SCORPIO was able to prognosticate overall survival for non-ICI patients in some, but not all, cancer types (Supplementary Fig. 16). However, in contrast to the ICI treatment context, its prognostic accuracy decreased for specific cancers such as bladder cancer (P = 0.2713), hepatobiliary cancer (P = 0.1038), esophageal cancer (P = 0.8886) and ovarian cancer (P = 0.4305; Fig. 3a and Supplementary Figs. 10a, 13a and 16). These findings suggest that SCORPIO is more effective at prognosticating overall survival in the context of ICI treatment. To understand how each feature contributes to SCORPIO's risk score prediction, we analyzed the relative effect of its 33 features in the training set using the SHapley Additive exPlanations (SHAP) approach. SHAP quantified the contribution of each feature to patient-to-patient variation in ICI efficacy (Fig. 4a). The top five features contributing the most were chloride (CL), ALB, hemoglobin (HGB), ECOG-PS and eosinophil proportion among white blood cells (EOS%). Figure 4b-e show representative patients from the hold-out test set with different risk scores and clinical responses. Each feature's contribution varied in direction and magnitude based on its value and the values of other features, demonstrating the model's complexity in predicting ICI efficacy for each patient. Next, we investigated how the top five features and the predicted risk score reflect characteristics of the tumor microenvironment (TME). We gathered an additional cohort of 264 patients with NSCLC, with available bulk RNA-sequencing (RNA-seq), blood test values (performed on the date of, or no more than 30 days before, the tumor biopsy) and clinical data. Using the Danaher signature, which was validated as the most accurate immune cell deconvolution method for NSCLC, we deconvoluted 14 immune cell types. We then analyzed the correlations between their abundances and the levels of the top five features, as well as the predicted risk score (Fig. 4f). Our findings showed that higher ALB levels were associated with increased abundances of mast cells, T cells, B cells, CD45 cells and regulatory T cells. Conversely, lower ECOG-PS was linked to greater abundances of T cells, B cells, CD45 cells, exhausted CD8 cells and cytotoxic cells. Additionally, a lower predicted risk score (indicating better-predicted response to immunotherapy) corresponded with higher abundances of mast cells, T cells, B cells, CD45 cells, regulatory T cells, natural killer CD56 dim cells and Th1 cells. We further analyzed the association between the abundances of the 14 immune cell types and the levels of the top 5 features, as well as the predicted risk score, in patients with head and neck (H&N) cancer (n = 32) from the MSK-I cohort (Fig. 4g). Compared to the NSCLC cohort, there were fewer significant associations, likely due to the smaller sample size. However, very similar relationships were observed -- various immune cell types were positively correlated with ALB levels, whereas ECOG-PS and predicted risk score were negatively correlated with many immune cell types. These results suggest that some features in SCORPIO reflect the TME status and a low predicted risk score corresponds to an immune-inflamed phenotype in patients. We also assessed whether the top five features correlated with TMB. Using patients from multiple MSKCC cohorts (n = 2,969), we found that TMB was generally not associated with the top five features or the risk scores, except in a few cancer types (Supplementary Fig. 17). Among the clinical trial cohorts, SCORPIO achieved its highest performance in prognosticating overall survival at 6, 12, 18, 24 and 30 months in the IMvigor211 trial (bladder cancer) with a median AUC(t) of 0.782, and in predicting clinical benefit in IMspire150 trial (melanoma), with an AUC of 0.684 (Fig. 2). In each clinical trial cohort, the three risk groups showed significantly different overall survival rates (P < 0.0001 for each trial; Fig. 5a). Similarly, clinical benefit rates varied significantly across risk groups (P values: 0.043 for IMpower133, 0.0027 for IMpower131 (ACNP), 0.001 for IMpower131 (ACP), 0.0004 for IMpower150 (ABCP), 0.0003 for IMspire150 and OAK, 0.0001 for IMpower150 (ACP) and <0.0001 for the remaining trials; Fig. 5b). Importantly, these results were independent of sex, age and PD-L1 expression (Supplementary Fig. 18). In clinical trials, SCORPIO outperformed PD-L1 staining in predicting clinical benefit and overall survival, as indicated by various performance metrics (Supplementary Fig. 19). To further test model generalizability, we analyzed a real-world cohort of patients treated at a large, comprehensive health system (MSHS), encompassing a diverse patient population. In this cohort, SCORPIO prognosticated overall survival at 6, 12, 18, 24 and 30 months following ICI with a median pan-cancer AUC(t) of 0.725 (Fig. 2), and the three risk groups had significantly different overall survival after ICI administration (Fig. 6). Across tumor types, HRs for death in the low-risk and moderate-risk groups compared to the high-risk group were 0.25 (95% CI, 0.18-0.34) and 0.41 (95% CI, 0.33-0.50), respectively. Importantly, all these results were independent of the line of therapy in which ICI was administered, sex, age and ECOG-PS (Supplementary Fig. 20). SCORPIO performed better in prognosticating overall survival in real-world cohorts compared to phase 3 clinical trials across most cancer types (Fig. 2). For example, in bladder cancer, the median AUC(t) in real-world cohorts was 0.809 (across all time points and cohorts), outperforming the 0.782 observed in the IMvigor211 trial. Similarly, for hepatobiliary cancer, the median AUC(t) in real-world data reached 0.746, surpassing the 0.704 reported in the IMbrave150 trial. Notably, SCORPIO demonstrated the strongest performance in RCC from the real-world cohorts, with a median AUC(t) of 0.829, higher than the 0.668 observed in the IMmotion151 trial. The model performed better in real-world cohorts, likely due to the broader range of patient characteristics, cancer types and treatment environments in the training data. This also suggests that the model effectively captures the complexities and variations found in everyday clinical practice, enhancing its applicability for predicting the efficacy of ICIs in diverse patient populations. Notably, the model performed better at prognosticating overall survival than predicting clinical benefit across most cancer types and cohorts, likely reflecting the robustness of overall survival as a reliable clinical endpoint, which is often prioritized in oncology for its clear and objective outcomes compared to clinical benefit. Furthermore, our analysis revealed that SCORPIO's performance in predicting clinical benefit is not uniform across different cancer types (Fig. 2). To understand the variability in performance across cancer types, we compared SHAP values between cancer-type-specific models and SCORPIO. This analysis revealed key features that SCORPIO's pan-cancer modeling approach may have overlooked. Our findings showed some variability in the importance of specific features in different cancer types within the SCORPIO model (Supplementary Fig. 21). For example, although SHAP analyses indicated that ALB and HGB were important in SCORPIO, their importance was reduced in cancer-type-specific models, particularly in bladder cancer, ovarian cancer, H&N cancer and NSCLC. Additionally, features like viral infection, relevant in H&N cancer due to human papillomavirus status, and platelet count, influential in both H&N cancer and melanoma, highlight the possibly unique biological characteristics of each cancer type. These variations may suggest that SCORPIO's pan-cancer approach may not fully capture the cancer-type-specific importance of certain features. Nevertheless, SCORPIO outperformed cancer-type-specific models in predicting overall survival and clinical benefit, demonstrating its robustness and generalizability (Supplementary Fig. 8). Targeted refinements that incorporate cancer-specific features may further enhance SCORPIO's accuracy in predicting clinical benefit, balancing the need for generalizability with the precision required for specific cancer types in the future.
Share
Share
Copy Link
Researchers develop an AI model called SCORPIO that uses routine blood tests to predict cancer patients' response to immunotherapy, potentially improving treatment decisions and accessibility.

AI Model SCORPIO Revolutionizes Cancer Immunotherapy Prediction
Researchers from Memorial Sloan Kettering Cancer Center (MSK) and the Tisch Cancer Institute at Mount Sinai have developed a groundbreaking artificial intelligence tool called SCORPIO that could transform cancer treatment decisions worldwide
1
2
. This innovative model uses routine blood tests and clinical data to predict patient responses to immune checkpoint inhibitors, a type of immunotherapy.Advantages Over Current Methods
SCORPIO offers several advantages over existing prediction methods:
-
Accessibility: Unlike current FDA-approved biomarkers that require tumor samples and expensive genomic testing, SCORPIO relies on widely available clinical data and routine blood tests
1
2
. -
Improved Accuracy: The model outperforms currently used tests in predicting patient outcomes
1
2
3
. -
Cost-Effectiveness: By using readily available data, SCORPIO could reduce healthcare costs and improve access to care
1
2
.
Development and Validation
The SCORPIO model was developed using a comprehensive approach:
-
Initial Development: Data from over 2,000 MSK patients across 17 cancer types was used to train the model
1
2
3
. -
Extensive Testing: The model was validated using:
- 2,100 additional MSK patients
- Nearly 4,500 patients from 10 global phase 3 clinical trials
- 1,200 patients from Mount Sinai
1
2
3
-
Diverse Dataset: In total, the study included nearly 10,000 patients across 21 different cancer types, representing the largest dataset in cancer immunotherapy to date
1
2
3
.
Technical Approach
SCORPIO employs ensemble machine learning, combining several tools to identify patterns in clinical data from blood tests and treatment outcomes
1
2
. The model uses data from complete blood count and comprehensive metabolic profile tests, which are routinely performed in clinics worldwide1
2
3
.Related Stories
Potential Impact
Dr. Luc Morris, co-senior author of the study, emphasizes the importance of patient selection for immunotherapy: "Immune checkpoint inhibitors are a very powerful tool against cancer, but they don't yet work for most patients. These drugs are expensive, and they can come with serious side effects"
1
2
3
.By improving prediction accuracy, SCORPIO could help:
- Enhance treatment decisions
- Reduce unnecessary side effects
- Lower healthcare costs
- Increase equitable access to care
1
2
3
Future Steps
The research team plans to:
- Collaborate with hospitals and cancer centers globally to further test and optimize the model
1
2
3
. - Develop a user-friendly interface for clinicians to access the tool easily
1
2
3
.
As this AI-driven approach continues to evolve, it has the potential to significantly improve cancer care by enabling more personalized and effective treatment strategies for patients worldwide.
References
Summarized by
Navi
[1]
Related Stories
Recent Highlights
1
OpenAI Releases GPT-5.4, New AI Model Built for Agents and Professional Work
Technology

2
Pentagon's Anthropic showdown exposes who controls AI guardrails in military contracts
Policy and Regulation

3
Anthropic challenges Pentagon supply chain risk label in court over AI usage restrictions
Policy and Regulation