OpenFold3 Emerges as Open-Source Alternative to AlphaFold3, Democratizing Protein Structure Prediction
2 Sources
2 Sources
[1]
Open-source protein structure AI aims to match AlphaFold
Scientists today released a 'sneak preview' of a new open-source artificial intelligence (AI) model that predicts 3D structures of proteins, and say it is close to matching the performance of Google DeepMind's revolutionary protein-folding AI AlphaFold3. The system, called OpenFold3, was developed by the OpenFold Consortium, a non-profit collaboration of academic and private research groups, headquartered in Davis, California. OpenFold3 uses proteins' amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA. The tool still doesn't have the same functionality as AlphaFold3, but "we wanted to get something out to the community as soon as possible", says Woody Sherman, the consortium's executive committee chair and chief innovation officer at the firm Psivant Therapeutics in Boston, Massachusetts. The team hopes to use researcher feedback after the preview release to improve the model. The system was trained on more than 300,000 molecular structures and a synthetic database of more than 40 million structures. Developing it has cost US$ 17 million so far. Unlike AlphaFold3, which is available for restricted academic use, any researcher or pharmaceutical company can use OpenFold3. "It's a big step forward in terms of the democratization of AI structural-biology tools," says Sherman. The preview release shares OpenFold3's code with researchers and allows them to start using and testing it. Sherman says that the consortium team is still working on technical improvements to the system and is planning a full release in the coming months. "We're close -- and we think, with this last little bit of effort, we're going to get fully to parity," he says. "This is a way for folks to get a flavour of OpenFold3 and start integrating it into their workflows and start building tools and start providing feedback." "I'm excited to test [OpenFold3] and see how [it] compares to existing models," says Stephanie Wankowicz, a computational structural biologist at Vanderbilt University in Nashville, Tennessee. OpenFold3 is part of a wider push to develop open-source versions of AlphaFold3. These efforts began in May 2024, when London-based Google DeepMind launched AlphaFold3 without initially sharing its underlying code. Researchers criticized the private firm's decision -- and later, in November 2024, DeepMind made the AlphaFold3 code and model weights, or all the trainable parameters in an AI model, available to academics (they remain unavailable for commercial use). Late last year, AI researcher Regina Barzilay, at the Massachusetts Institute of Technology in Cambridge, and her colleagues released a similar open-access model called Boltz. A second of version of the system, released earlier this year, predicts protein structures and estimates how strongly they bind to small ligands. "It is great to see more open-source tools for modelling biomolecular interactions," says Barzilay. "While AlphaFold3 marked a major scientific breakthrough, fully open-source replications, such as Boltz-1, have already delivered substantial benefits -- both as ready-to-use resources and as powerful foundations for fine-tuning on specialized data sets. OpenFold3 will provide additional options for the research community." Wankowicz says that comparing AlphaFold3, OpenFold3 and Boltz will help researchers to better tease out "the plethora of puzzle pieces that make the algorithms work" and "which puzzle piece is most important for different functions of the algorithm". Several pharmaceutical and biotechnology firms have already committed to using OpenFold3 to design drugs for autoimmune disorders and cell therapies, as well as developing molecules that protect plants and crops. Sherman says the tool's full release will be only the start. The consortium plans to continue updating the model and will expand the training data to include more molecules, such as water. "We are already starting to work on predicting binding affinity and building in other properties of proteins like thermal stability," he adds.
[2]
The AI model OpenFold3 takes a crucial step in making protein predictions
Scientists open up AlphaFold3's secrets with the new OpenFold3 model A new AI model is opening the black box of the leading artificial intelligence tool for predicting how proteins will interact with small molecules, such as drugs. The model, OpenFold3, which launched October 28, is a reconstruction of Google DeepMind's AlphaFold3. A large consortium of researchers led by Mohammed AlQuraishi at Columbia University painstakingly dissected AlphaFold3's code and created a facsimile of the AI platform, which predicts the structure of proteins paired with other molecules, including nucleic acids and chemicals in drugs. AlphaFold3 can only be used by individuals, non-commercial organizations or journalists. But companies -- and anyone else -- can use the open-source OpenFold3 model for commercial purposes, including drug development. Predicting protein-molecule pairings is important in designing drugs "because this is how biology works. Biology is not proteins in isolation. It's biomolecules interacting with each other," says Woody Sherman, founder and chief innovation officer at Boston-based Psivant Therapeutics. Sherman also chairs the OpenFold executive committee. Proteins are some of the hardest working molecules in the body. How these workhorses perform depends largely on their shape. AlphaFold2 cracked the problem of predicting what shapes proteins will adopt. The team behind the AI model shared in the 2024 Nobel Prize in chemistry for the achievement. AlphaFold3 introduced interactions with other proteins and molecules to the mix. But unlike AlphaFold2, DeepMind didn't initially open the AlphaFold3 code for other researchers to explore, at least not until hundreds of scientists signed a petition calling for transparency. "It's hard to evaluate a computational product without seeing the raw information," says Stephanie Wankowicz, a computational structural biologist at Vanderbilt University who coauthored the petition. It's necessary for other researchers to have the code to test accuracy and reliability of the predictions and to learn what other data are necessary to make the model better, Wankowicz says. Re-creating AlphaFold2 gave OpenFold creators insight into how the AI works, she says. AlphaFold2 was billed as an AI model that learns how proteins fold based on their amino acid building blocks, but it actually memorizes protein structures it has seen before and uses those memories to predict how similar proteins may appear, Wankowicz says. Looking under AlphaFold3's hood may yield similar insights into protein-drug pairs. Other teams have tried to reproduce AlphaFold3 and "have gotten close, but not super precise," Wankowicz says. That's because it is difficult to reproduce subtle tricks and tweaks that are in the AlphaFold3 creators' heads but don't appear in the code or supplemental information, Sherman says. Some are technical settings used for certain parts of the calculation. "Nobody's specifying that," he says. "But details matter, especially when you're dealing with the large models and with lots of data." The OpenFold3 team did its best to replicate AlphaFold3, he says, but some differences remain. Biology also matters, Sherman says. In cells, proteins are bathed in water and ions. They vibrate and move. None of that is captured in the static images created by AI models or by lab-made snapshots of crystalized proteins. The OpenFold3 team hopes to add water and dynamic movement into its model to better reflect how proteins exist in nature, Sherman says. Even before its official release OpenFold3 was embraced by pharmaceutical companies. Five companies banded together in the Federated OpenFold3 Initiative to train the AI model on proprietary data and build a more powerful prediction tool while still keeping company secrets. That partnership was announced October 1 by Apheris, a Berlin-based company that runs the group platform. Only about 2 percent of the protein structures in publicly available databases on which AlphaFold3 and OpenFold3 were trained are paired with molecules that have druglike properties, says Robin Röhm, cofounder and chief executive of Apheris. Drug companies have thousands of such structures in their databases. Each company in the federation will train a version of OpenFold3 on about 4,000 to 8,000 protein-drug pairs in its own library, Röhm says. Apheris aggregates those locally trained AIs into a centralized version that has the knowledge about how about 20,000 proteins and drugs interact but doesn't contain the proprietary data. The global version goes back to each company for another round of training and so on. Despite the expanded datasets, don't expect dramatic changes yet in drug discovery, Sherman says. OpenFold3 "is a starting point," he says. "It's going to be the next stage, and the next stage and the next stage that are where we're really going to start seeing that meaningful impact on drug discovery."
Share
Share
Copy Link
Scientists release OpenFold3, an open-source AI model that aims to match AlphaFold3's protein structure prediction capabilities. The $17 million project offers unrestricted access for both academic and commercial use, addressing transparency concerns in AI-driven drug discovery.
OpenFold3 Launch Addresses Scientific Community's Call for Transparency
The OpenFold Consortium has released a preview of OpenFold3, an open-source artificial intelligence model designed to predict three-dimensional protein structures and their interactions with other molecules. The system represents a significant step toward democratizing access to advanced protein-folding AI technology, addressing longstanding concerns about the restricted availability of Google DeepMind's AlphaFold3
1
.Developed by a non-profit collaboration of academic and private research groups headquartered in Davis, California, OpenFold3 has cost $17 million to develop and was trained on more than 300,000 molecular structures plus a synthetic database of over 40 million structures. Unlike AlphaFold3, which is available only for restricted academic use, OpenFold3 can be used by any researcher or pharmaceutical company without commercial limitations
1
.Technical Capabilities and Development Approach
OpenFold3 uses proteins' amino acid sequences to map their 3D structures and model interactions with other molecules, including drugs and DNA. The consortium's executive committee chair, Woody Sherman, who also serves as chief innovation officer at Psivant Therapeutics, emphasized the team's commitment to community access: "We wanted to get something out to the community as soon as possible"
1
.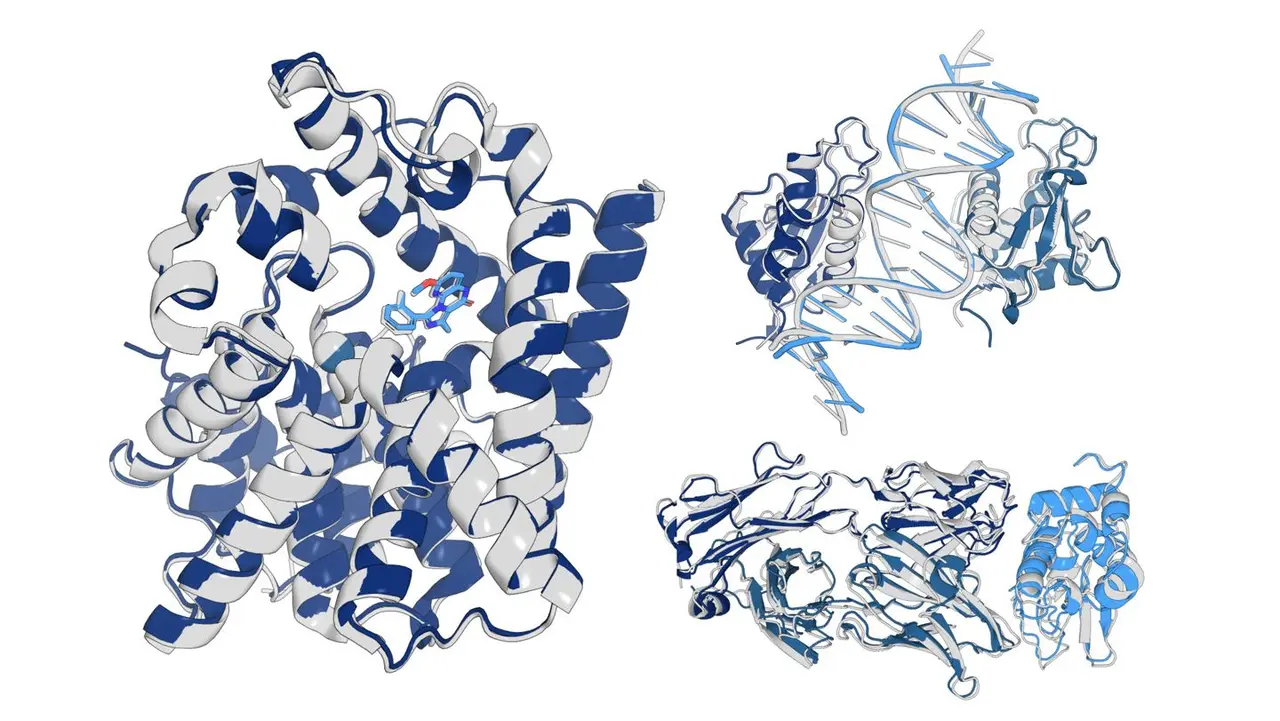
Source: Science News
The model represents a painstaking reconstruction of AlphaFold3's capabilities. Led by Mohammed AlQuraishi at Columbia University, researchers dissected AlphaFold3's code to create this facsimile, though some technical differences remain due to undocumented implementation details
2
.Addressing Scientific Transparency Concerns
The development of OpenFold3 emerged from widespread criticism within the scientific community regarding AlphaFold3's initial lack of code transparency. When Google DeepMind launched AlphaFold3 in May 2024 without sharing its underlying code, hundreds of scientists signed a petition calling for transparency. Stephanie Wankowicz, a computational structural biologist at Vanderbilt University who coauthored the petition, explained: "It's hard to evaluate a computational product without seeing the raw information"
2
.DeepMind eventually made AlphaFold3's code and model weights available to academics in November 2024, though they remain unavailable for commercial use. The open-source movement has gained momentum with other similar projects, including Regina Barzilay's Boltz model from MIT, which was released in late 2024
1
.Related Stories
Commercial Applications and Industry Adoption
Several pharmaceutical and biotechnology firms have already committed to using OpenFold3 for various applications, including designing drugs for autoimmune disorders, developing cell therapies, and creating molecules for plant and crop protection. The commercial viability of OpenFold3 has attracted significant industry interest, with five companies forming the Federated OpenFold3 Initiative to train the AI model on proprietary data while maintaining confidentiality
2
.Robin Röhm, cofounder and CEO of Berlin-based Apheris, which runs the federation platform, noted that only about 2 percent of publicly available protein structures are paired with druglike molecules, while pharmaceutical companies possess thousands of such structures in their databases. The federation allows companies to train OpenFold3 on 4,000 to 8,000 protein-drug pairs from their libraries, creating a more powerful prediction tool without exposing proprietary information
2
.References
Summarized by
Navi
Related Stories
Google DeepMind Open-Sources AlphaFold 3: A Game-Changer for Drug Discovery and Molecular Biology
11 Nov 2024•Science and Research

AlphaFold Upgrade: AI Now Predicts Large Protein Structures and Integrates Experimental Data
05 Nov 2024•Science and Research

MassiveFold: Revolutionizing Protein Structure Prediction with Optimized AlphaFold
14 Nov 2024•Science and Research

Recent Highlights
1
OpenAI Releases GPT-5.4, New AI Model Built for Agents and Professional Work
Technology

2
Anthropic takes Pentagon to court over unprecedented supply chain risk designation
Policy and Regulation

3
Meta smart glasses face lawsuit and UK probe after workers watched intimate user footage
Policy and Regulation





