AI Breakthrough Accelerates Discovery of New Tuberculosis Drug Candidates
2 Sources
2 Sources
[1]
AI-based technology accelerates discovery of new tuberculosis drug candidates
University of California - San DiegoFeb 7 2025 Tuberculosis is a serious global health threat that infected more than 10 million people in 2022. Spread through the air and into the lungs, the pathogen that causes "TB" can lead to chronic cough, chest pains, fatigue, fever and weight loss. While infections are more extensive in other parts of the world, a serious tuberculosis outbreak currently unfolding in Kansas has led to two deaths and has become one of the largest on record in the United States. While tuberculosis is typically treated with antibiotics, the rise of drug-resistant strains has led to an urgent need for new drug candidates. A new study published in the Proceedings of the National Academy of Sciences describes the novel use of artificial intelligence to screen for antimicrobial compound candidates that could be developed into new tuberculosis drug treatments. The study was led by researchers at the University of California San Diego, Linnaeus Bioscience Inc. and the Center for Global Infectious Disease Research at the Seattle Children's Research Institute. Linnaeus Bioscience is a San Diego-based biotechnology company founded on technology developed in the UC San Diego School of Biological Sciences laboratories of Professor Joe Pogliano and Dean Kit Pogliano. Their bacterial cytological profiling (BCP) method provides a shortcut for understanding how antibiotics function by rapidly determining their underlying mechanisms. The search for new tuberculosis drug targets under traditional laboratory methods has historically proven to be arduous and time-consuming due in part to the difficulty of understanding how new drugs work against Mycobacterium tuberculosis, the bacterium that causes the disease. The new PNAS study describes the development of "MycoBCP," a next-generation technology developed with funding from the Gates Foundation. The new method adapts BCP with deep learning - a type of artificial intelligence that uses brain-like neural networks - to overcome traditional challenges and open new views of Mycobacterium tuberculosis cells. This is the first time that this kind of image analysis using machine learning and AI has been applied in this way to bacteria. Tuberculosis images are inherently difficult to interpret by the human eye and traditional lab measurements. Machine learning is much more sensitive in being able to pick up the differences in shapes and patterns that are important for revealing underlying mechanisms." Joe Pogliano, paper co-author, professor in the Department of Molecular Biology Over two years in development, study lead authors Diana Quach and Joseph Sugie shaped the MycoBCP technology by training AI tools known as convolutional neural networks with more than 46,000 images of TB cells (now at Linnaeus Bioscience, Quach and Sugie both received their PhDs from the Shu Chien-Gene Lay Department of Bioengineering and completed postdoctoral appointments in the Pogliano labs in the Department of Molecular Biology). "Tuberculosis cells are clumpy and seem to always stick close to each other, so defining cell boundaries didn't seem possible," said Sugie, chief technology officer at Linnaeus Bioscience. "Instead, we jumped straight into letting the computer analyze the patterns in the images for us." Linnaeus teamed up with tuberculosis expert Tanya Parish of Seattle Children's Research Institute to develop BCP for mycobacteria. The new system has already vastly accelerated the team's TB research capabilities and helped identify optimal candidate compounds for drug development. "A critical component of progressing towards new drug candidates is defining how they work, which has been technically challenging and takes time," said Parish, a co-author of the study. "This technology expands and accelerates our ability to do this and allows us to prioritize which molecules to work on based on their mode of action. We were excited to collaborate with Linnaeus in their work to develop this technology to M. tuberculosis." UC San Diego biotech spinoff tackles global health issue Linnaeus Bioscience was launched in 2012 with a UC San Diego-developed technology that promised to change the face of understanding how antibiotics work. "We developed bacterial cytological profiling and it allowed us to look at bacterial cells in a new way," said Joe Pogliano. "It allowed us to really see how cells look after treatment with antibiotics so we could interpret their underlying mechanisms. We describe this method as equivalent to performing an autopsy on a bacterial cell." Establishing Linnaeus Bioscience in the regional San Diego biotechnology hub allowed Joe and Kit Pogliano to push the BCP technology out into the marketplace, where other companies could have access to it. The company now receives samples from all over the world for rapid analysis and identification of new bacterial drug candidates. Pogliano credits the biotechnology community, especially the company's early home in the San Diego JLABS incubator, which supports early stage biotech companies, as critical to the company's growth and success. "We could not have gotten Linnaeus Bioscience off the ground if not for the supportive biotech community and the infrastructure provided at JLABS," said Pogliano. "All of the company's employees at Linnaeus obtained their PhDs at UC San Diego so this has become a great UC San Diego research, alumni and San Diego biotech community success story, culminating in this new AI platform to help solve the antibiotic resistance crisis." In addition to Quach, Pogliano and Sugie, coauthors of the paper include Marc Sharp, Sara Ahmed, Lauren Ames, Amala Bhagwat, Aditi Deshpande and Tanya Parish. University of California - San Diego Journal reference: Quach, D., et al. (2025). Deep learning-driven bacterial cytological profiling to determine antimicrobial mechanisms in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2419813122.
[2]
AI Accelerates the Search for New Tuberculosis Drug Targets | Newswise
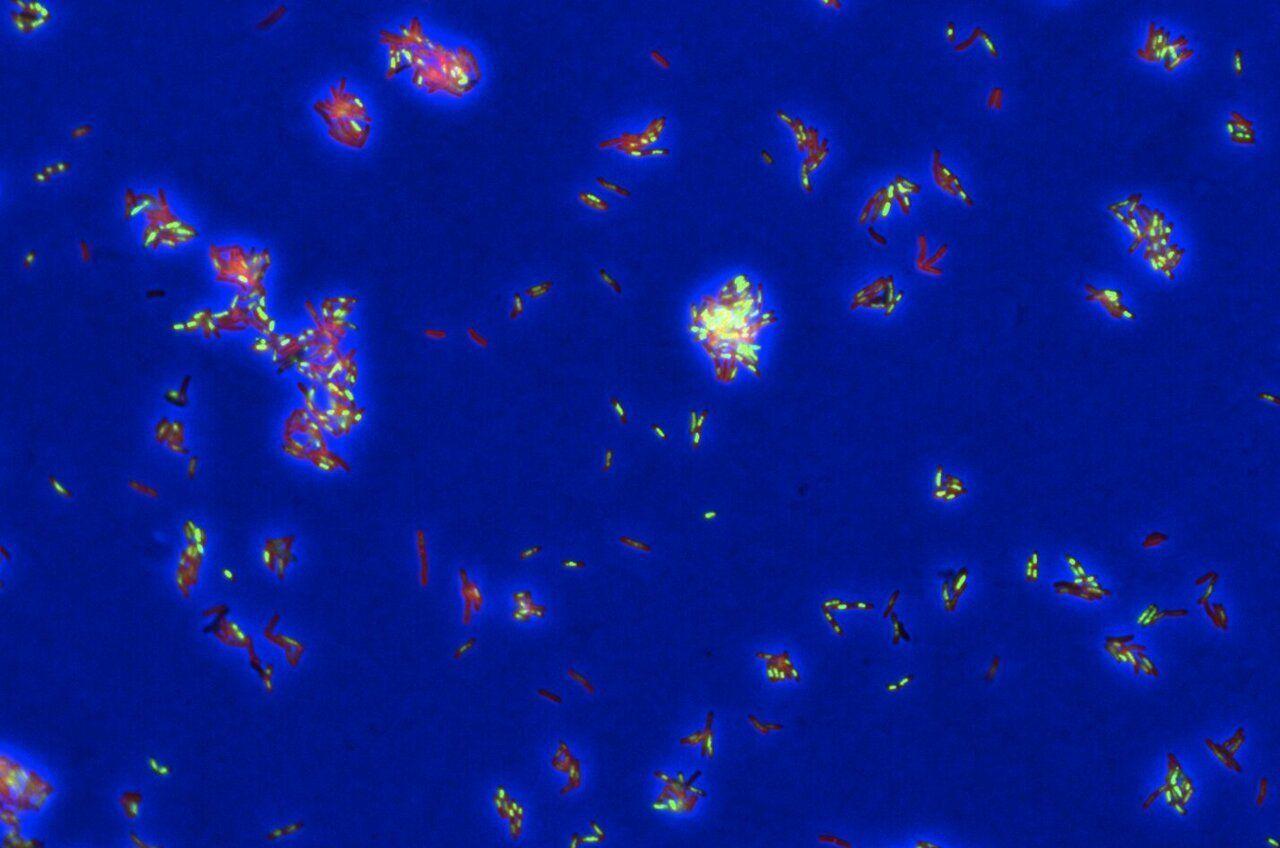
A fluorescence microscopy image reveals the tuberculosis-causing bacterium Mycobacterium tuberculosis after an antimicrobial treatment. Membranes are stained red, DNA blue and areas of membrane permeability appear green. These dramatic changes in bacterial cell structure form consistent patterns that can be used to identify how potential new treatments work -- a critical step in developing effective therapies for the globally significant tuberculosis disease. Tuberculosis is a serious global health threat that infected more than 10 million people in 2022. Spread through the air and into the lungs, the pathogen that causes "TB" can lead to chronic cough, chest pains, fatigue, fever and weight loss. While infections are more extensive in other parts of the world, a serious tuberculosis outbreak currently unfolding in Kansas has led to two deaths and has become one of the largest on record in the United States. While tuberculosis is typically treated with antibiotics, the rise of drug-resistant strains has led to an urgent need for new drug candidates. A new study published in the Proceedings of the National Academy of Sciences describes the novel use of artificial intelligence to screen for antimicrobial compound candidates that could be developed into new tuberculosis drug treatments. The study was led by researchers at the University of California San Diego, Linnaeus Bioscience Inc. and the Center for Global Infectious Disease Research at the Seattle Children's Research Institute. Linnaeus Bioscience is a San Diego-based biotechnology company founded on technology developed in the UC San Diego School of Biological Sciences laboratories of Professor Joe Pogliano and Dean Kit Pogliano. Their bacterial cytological profiling (BCP) method provides a shortcut for understanding how antibiotics function by rapidly determining their underlying mechanisms. The search for new tuberculosis drug targets under traditional laboratory methods has historically proven to be arduous and time-consuming due in part to the difficulty of understanding how new drugs work against Mycobacterium tuberculosis, the bacterium that causes the disease. The new PNAS study describes the development of "MycoBCP," a next-generation technology developed with funding from the Gates Foundation. The new method adapts BCP with deep learning -- a type of artificial intelligence that uses brain-like neural networks -- to overcome traditional challenges and open new views of Mycobacterium tuberculosis cells. "This is the first time that this kind of image analysis using machine learning and AI has been applied in this way to bacteria," said paper co-author Joe Pogliano, a professor in the Department of Molecular Biology. "Tuberculosis images are inherently difficult to interpret by the human eye and traditional lab measurements. Machine learning is much more sensitive in being able to pick up the differences in shapes and patterns that are important for revealing underlying mechanisms." Over two years in development, study lead authors Diana Quach and Joseph Sugie shaped the MycoBCP technology by training AI tools known as convolutional neural networks with more than 46,000 images of TB cells (now at Linnaeus Bioscience, Quach and Sugie both received their PhDs from the Shu Chien-Gene Lay Department of Bioengineering and completed postdoctoral appointments in the Pogliano labs in the Department of Molecular Biology). "Tuberculosis cells are clumpy and seem to always stick close to each other, so defining cell boundaries didn't seem possible," said Sugie, chief technology officer at Linnaeus Bioscience. "Instead, we jumped straight into letting the computer analyze the patterns in the images for us." Linnaeus teamed up with tuberculosis expert Tanya Parish of Seattle Children's Research Institute to develop BCP for mycobacteria. The new system has already vastly accelerated the team's TB research capabilities and helped identify optimal candidate compounds for drug development. "A critical component of progressing towards new drug candidates is defining how they work, which has been technically challenging and takes time," said Parish, a co-author of the study. "This technology expands and accelerates our ability to do this and allows us to prioritize which molecules to work on based on their mode of action. We were excited to collaborate with Linnaeus in their work to develop this technology to M. tuberculosis." UC San Diego biotech spinoff tackles global health issue Linnaeus Bioscience was launched in 2012 with a UC San Diego-developed technology that promised to change the face of understanding how antibiotics work. "We developed bacterial cytological profiling and it allowed us to look at bacterial cells in a new way," said Joe Pogliano. "It allowed us to really see how cells look after treatment with antibiotics so we could interpret their underlying mechanisms. We describe this method as equivalent to performing an autopsy on a bacterial cell." Establishing Linnaeus Bioscience in the regional San Diego biotechnology hub allowed Joe and Kit Pogliano to push the BCP technology out into the marketplace, where other companies could have access to it. The company now receives samples from all over the world for rapid analysis and identification of new bacterial drug candidates. Pogliano credits the biotechnology community, especially the company's early home in the San Diego JLABS incubator, which supports early stage biotech companies, as critical to the company's growth and success. "We could not have gotten Linnaeus Bioscience off the ground if not for the supportive biotech community and the infrastructure provided at JLABS," said Pogliano. "All of the company's employees at Linnaeus obtained their PhDs at UC San Diego so this has become a great UC San Diego research, alumni and San Diego biotech community success story, culminating in this new AI platform to help solve the antibiotic resistance crisis." In addition to Quach, Pogliano and Sugie, coauthors of the paper include Marc Sharp, Sara Ahmed, Lauren Ames, Amala Bhagwat, Aditi Deshpande and Tanya Parish. The research funding was provided by the Bill and Melinda Gates Foundation (INV-040479).
Share
Share
Copy Link
Researchers at UC San Diego and partners have developed an AI-powered technology called MycoBCP that significantly speeds up the identification of potential new tuberculosis treatments, addressing the urgent need for solutions against drug-resistant strains.

AI-Powered Technology Revolutionizes Tuberculosis Drug Discovery
In a groundbreaking development, researchers have harnessed the power of artificial intelligence to accelerate the discovery of new tuberculosis drug candidates. A study published in the Proceedings of the National Academy of Sciences details the novel use of AI in screening antimicrobial compounds that could lead to new treatments for tuberculosis (TB), a disease that infected over 10 million people in 2022
1
2
.The Urgent Need for New TB Treatments
Tuberculosis remains a serious global health threat, with drug-resistant strains posing a particular challenge. The urgency of finding new treatments is underscored by a recent outbreak in Kansas, which has become one of the largest on record in the United States, resulting in two deaths
1
2
.MycoBCP: A Next-Generation AI Solution
The study introduces "MycoBCP," a next-generation technology developed with funding from the Gates Foundation. This innovative method combines bacterial cytological profiling (BCP) with deep learning to overcome traditional challenges in understanding how new drugs work against Mycobacterium tuberculosis, the bacterium causing TB
1
.AI's Unique Advantage in TB Research
Dr. Joe Pogliano, a co-author of the study and professor at UC San Diego, explains the significance of this approach: "This is the first time that this kind of image analysis using machine learning and AI has been applied in this way to bacteria. Machine learning is much more sensitive in being able to pick up the differences in shapes and patterns that are important for revealing underlying mechanisms"
1
.Development and Training of AI Tools
Lead authors Diana Quach and Joseph Sugie developed the MycoBCP technology over two years, training convolutional neural networks with over 46,000 images of TB cells. This approach overcame the challenge of analyzing clumpy tuberculosis cells, which are difficult to interpret using traditional methods
1
2
.Related Stories
Collaboration and Impact
Linnaeus Bioscience, a San Diego-based biotechnology company, collaborated with tuberculosis expert Tanya Parish of Seattle Children's Research Institute to develop BCP for mycobacteria. The new system has already accelerated TB research capabilities and helped identify optimal candidate compounds for drug development
1
2
.From University Research to Biotech Success
Linnaeus Bioscience, founded in 2012 based on technology developed at UC San Diego, has played a crucial role in bringing this innovation to the market. The company's success story highlights the importance of supportive biotech communities and infrastructure, such as the San Diego JLABS incubator, in fostering groundbreaking research and its commercial applications
1
2
.This AI-driven approach to tuberculosis drug discovery represents a significant leap forward in the fight against a persistent global health threat, offering hope for more effective treatments in the near future.
References
Summarized by
Navi
Related Stories
AI-Powered Tool DECIPHAER Unveils Molecular Mechanisms of Tuberculosis Drug Action
26 Aug 2025•Health
AI Accelerates Antibiotic Discovery: New Drug Targets IBD with Unprecedented Precision
03 Oct 2025•Health

AI Model Predicts Antibiotic Resistance in Bacteria with High Accuracy
03 Apr 2025•Science and Research
Recent Highlights
1
Pentagon threatens to cut Anthropic's $200M contract over AI safety restrictions in military ops
Policy and Regulation

2
ByteDance's Seedance 2.0 AI video generator triggers copyright infringement battle with Hollywood
Policy and Regulation

3
OpenAI closes in on $100 billion funding round with $850 billion valuation as spending plans shift
Business and Economy





