Google DeepMind's AlphaGenome AI tackles the 98% of human genome that doesn't code for proteins
14 Sources
14 Sources
[1]
How DeepMind's genome AI could help solve rare disease mysteries
When more than 100 researchers voluntarily locked themselves in a room last year to tackle some of the hardest conditions in medicine, they turned to artificial intelligence. As part of an effort, called the Undiagnosed Hackathon, to crack 29 undiagnosed conditions researchers deployed AlphaGenome -- an AI model developed by Google DeepMind in London that was described in Nature on 28 January. AlphaGenome -- an AI tool that was made available to scientists last year -- can predict the diverse effects of mutations in non-coding DNA sequences, including how they might affect the activity of nearby genes. Deciphering the 98% of the human genome that does not code for proteins is one of biology's grand challenges. Mutations in these sequences are especially vexing to researchers seeking to uncover the genetic basis for rare, often fatal diseases. "These are variants that, to be quite honest, often get triaged," says Eric Klee, a bioinformatician at the Mayo Clinic in Rochester, Minnesota, who co-led the Undiagnosed Hackathon in September last year. The three-day event at the Mayo Clinic in Rochester, as well as two previous hackathons in Europe, were organized by the Wilhelm Foundation -- a charity in Brottby, Sweden, that advocates for families affected by undiagnosed rare diseases. The charity was founded by Helene and Mikk Cederroth, who lost three of their four children to an undiagnosed disease, and named the foundation after their eldest son, who died at the age of 16. Around 350 million people have an undiagnosed rare condition, but only a fraction can be diagnosed using existing technologies such as genome sequencing. "If you don't have a diagnosis, you are left behind," says Helene Cederroth. Efforts to diagnose rare diseases tend to focus on mutations in protein-coding regions of the genome, known as the exome. To see if AlphaGenome could help to interpret the effects non-coding variants, Klee tested its prediction for a variant that he and his colleagues had linked to an individual's diagnosis, before the September 2025 hackathon. Experimental work showed that the mutation altered gene expression in cardiac cells, but not in neural cells, which was in line with the symptoms the individual experienced. AlphaGenome's predictions of the variant's effects supported this conclusion, Klee says. At the Mayo Clinic hackathon, researchers turned to AlphaGenome to investigate several conditions, Klee says. These tended to be in patients who carried one copy of a 'recessive' gene mutation that can cause disease if another mutation affects the other copy of the gene. The researchers used the AI tool to predict the effects of suspect non-coding variants, then see if they might lead to a disease. However, none of these predictions led to a diagnosis, Klee says. Of the 29 conditions that the Undiagnosed Hackathon took on, six were diagnosed at the event using other approaches, another has since been linked to a mutation and Klee is hopeful that further RNA expression data will soon confirm an eighth. The next Undiagnosed Hackathon will take place from 3 to 5 February in Hyderabad, India. Klee hopes that AlphaGenome and other AI tools will soon prove helpful in diagnosing rare diseases, when other approaches fail. As a result of the Hackathon, dozens of rare disease researchers now have experience with the tool. "We've opened up the door to 100-plus people from around the world on how you can think about using something like AlphaGenome," Klee says. At a previous Undiagnosed Hackathon, held in the Netherlands in 2024, researchers used a Google DeepMind model, called AlphaMissense, for deciphering the effects of protein sequence-altering mutations. The event help diagnose someone with a rare condition called Rothmund-Thomson syndrome, which affects the eyes, skin and other organ systems. Last year, a team of researchers used another AI model to predict the effect of such 'missense' mutations called popEVE to identify more than 100 potential disease-causing genes using genome data from people with developmental disorders. They are now collaborating with researchers in Senegal to help in the diagnosis of rare conditions. Google DeepMind has released the code and weights underlying AlphaGenome for non-commercial use, which could make it easier for researchers to customize the model to help diagnose rare disease. During the hackathon last year, researchers used the model to predict the effects of one specified variant at a time, but it would be more useful to assess numerous mutations at once, Klee says.
[2]
How AlphaGenome Is Changing the Genomic Research Landscape
The team at Google DeepMind behind that Nobel Prize-winning platform then turned their lens from from the structure of proteins to how these molecules function in the body. Applying similar machine-learning methods, they first developed AlphaMissense, an AI tool for predicting which changes in protein structure are likely to cause disease. AlphaProteo, a system for designing proteins that bind to specific molecular targets, came next. Now the architects of the Alpha platform are pushing beyond proteins into genomics, seeking to decipher how the vast regulatory regions of DNA shape when, where, and how genes are turned on and off. Enter AlphaGenome. Described as a "Swiss Army knife for exploring non-coding DNA," the deep-learning tool offers a way to systematically interpret the 98 percent of the genome that does not encode instructions for making proteins, but instead orchestrates how those genetic instructions are used inside the cell. "This allows us to model intricate processes... with unprecedented precision," Žiga Avsec, head of genomics at Google DeepMind, said in a press conference unveiling the new tool. AlphaGenome has its limitations. For instance, the tool's training data draw largely from bulk tissue datasets, curbing its reliability in rare cell types or specific developmental stages, notes Christina Leslie, a computational biologist at Memorial Sloan Kettering Cancer Center. "Generalization to new cell types is a huge limitation," she says. It also struggles to capture distant effects when regulatory regions are hundreds of thousands to millions of DNA letters away from their target genes, Leslie pointed out. Even so, the model is helping scientists to prioritize which genetic variants are most likely to matter, narrowing the search from across the genome to a manageable set of testable hypotheses. "It is the state of the art right now," Leslie says. According to DeepMind, thousands of scientists around the world are already using AlphaGenome, which is freely available on GitHub for academic research purposes. It is being put to work across a range of applications, including pinpointing genetic drivers of cancer and rare diseases, discovering new drug targets, and designing synthetic strands of DNA with tailored regulatory functions. "It's exciting to have things like AlphaGenome come out and perform much better than all the other dedicated algorithms that are exploring various aspects of genome biology," says Richard Young, a biologist at the Whitehead Institute for Biomedical Research who has collaborated with Google DeepMind on its AI co-scientist platform but was not involved in AlphaGenome. "It's a huge accelerator." The arrival of AlphaGenome marks another step in AI's steady advance into some of biology's most stubborn and consequential challenges. For DeepMind, there is also a clear industrial logic at work. The company's growing stable of biological models -- spanning protein structure, mutation, and generation, and now genomic regulation -- amounts to a vertically integrated platform for molecular prediction. That platform, in turn, should help unlock new diagnostic capabilities and therapeutic strategies, according to Pushmeet Kohli, vice president of science and strategic initiatives at Google DeepMind. "All these different models are solving key problems that are relevant for understanding biology," Kohli says. AlphaGenome is the latest -- and most expansive -- piece of that strategy. Trained on raw DNA, the model predicts 11 types of biological signals that help determine how genes are used inside cells. These include whether a gene is turned on or off, where gene activity begins, how genetic messages are edited, how tightly DNA is packed, which regulatory proteins bind to it, and how distant regions of the genome interact with one another. Many of these features already have their own specialty AI tools -- SpliceAI for splice site prediction, ChromBPNet for local chromatin accessibility, Orca for three-dimensional genome architecture. But such tools are typically used in isolation, requiring researchers to stitch together results from multiple sources. "AlphaGenome replaces this fragmentation with a more unified framework, which is more convenient and user-friendly -- and we hope this will accelerate scientists' workflows," says Natasha Latysheva, a computational geneticist at Google DeepMind. And while there have been attempts to capture all manner of regulatory effects in a single model, earlier architectures such as Borzoi and Enformer typically traded fine-scale resolution for breadth of biological coverage. AlphaGenome tries to escape that trade-off. The model can ingest up to one million DNA letters at a time, preserving long-range regulatory context, while still making predictions at single-base-pair resolution. In practical terms, that means it can ask how a change in one nucleotide might reverberate across a vast swathe of the genome. The new paper presents several demonstrations of this capability. In one case, AlphaGenome correctly predicted how a tiny deletion disrupts a splice site in a gene involved in blood vessel biology, leading to reduced RNA output. In another, it captured how mutations near a cancer-linked gene boost its activity, helping to drive an aggressive form of leukemia. Whether this predictive power generalizes beyond well-studied genes remains an open question, though. "This is obviously a potentially valuable tool -- but it's a tool," says Charles Mullighan, deputy director of the St. Jude Children's Research Comprehensive Cancer Center. "It's not a final point of discovery, but it's going to be a very important tool for giving insights that then might guide further functional analyses and experiments." One "quirk" of the system, notes Latysheva, is its bias toward false negatives over false positives, meaning it is more likely to miss a genuinely important DNA variant rather than incorrectly flag a harmless one. "But the flip side of that is if it does predict a strong effect, it's actually very accurate," she says. So, when the model serves up a strong prediction, "you can have a decent amount of confidence that it knows what it's doing." That confidence proved useful for Y-h. Taguchi and Kenta Kobayashi from Chuo University in Japan when they set out to stress-test a data-driven link between sleep deprivation and specific neuronal cell types. Early adopters of AlphaGenome, the bioinformatics researchers used the AI tool as an independent cross-check, confirming that genes implicated by sleep loss were especially active in their neurons of interest -- just as their earlier analysis of gene-expression data from brain tissue had predicted. "AlphaGenome succeeded in the cross validation," says Takuchi, who published the results January 1 in the journal Genes. That sort of validation underscores AlphaGenome's role. Like AlphaFold before it, the system is not meant to explain biology in full, but to make its most opaque regions easier to explore.
[3]
Google DeepMind unleashes new AI AlphaGenome to investigate DNA's 'dark matter'
DeepMind's AlphaGenome AI model could help solve the problem of predicting how variations in noncoding DNA shape gene expression DNA is the blueprint for life, influencing everything about us -- including our health. We know that our genes, the genetic "words" that encode proteins, play a major role in our wellbeing. But the vast majority of our genome -- more than 98 percent, in fact -- consists of DNA that doesn't build proteins. Once disregarded as "junk DNA," scientists now know that this molecular dark matter is crucial for determining gene activity in ways that keep us healthy -- or cause disease. Exactly how this mysterious DNA shapes gene expression is a mystery -- but now the AI lab Google DeepMind has built a model that DeepMind says can predict the function of long stretches of noncoding DNA. The information it turns up could help solve the problem of predicting how these chunks of DNA influence our health. Called AlphaGenome, the model takes in sequences of up to one million DNA letters, also known as base pairs, and predicts how mutations in those stretches affect gene expression. The model is described today in Nature. The tool, a version of which DeepMind has made freely available to other researchers, could help scientists narrow down theories for how certain DNA changes affect gene function. In turn, this knowledge could help scientists craft better treatments for genetic diseases. On supporting science journalism If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today. "Ever since the human genome was sequenced, people have been trying to understand the semantics of it -- this has been a longstanding goal for DeepMind," says Pushmeet Kohli, the company's vice president for science and a coauthor of the new study. "It's like you have a huge book of three billion characters and something wrong happened in this book." "AlphaGenome can be used to say, 'If you change these words, what would be the effect?'" he adds. AlphaGenome works by combining information from several datasets focused on different aspects of gene expression -- how genes are turned on or off. The model is a successor of sorts to DeepMind's AlphaFold, an AI model that predicts the structure of almost every known protein from its amino acid sequence -- a central problem in biology. The researchers behind that effort shared the Nobel Prize in Chemistry in 2024. And in 2023 DeepMind released AlphaMissense, another AI tool that predicts how mutations in the regions of the genome that do generate proteins affect gene function. According to DeepMind's researchers, AlphaGenome performs as well or better than most other methods they tested. Previous tools generally required a trade-off between the length of a DNA sequence that could be used as input and accuracy. A key advance of AlphaGenome's approach is the ability to make accurate predictions about the function of extremely long genome sequences. "The genome is like the recipe of life," Kohli said in a press briefing about the work. "And really understanding 'What is the effect of changing any part of the recipe?' is what AlphaGenome sort of looks at." AlphaGenome is a research tool -- it's not meant to be used clinically and its results can't be easily applied to individual humans. But it could have applications in understanding how the genome regulates genes in different types of cells or tissues. It could also help us understand diseases through massive genome-wide association studies or for studying cancer, since tumors can have many different genetic mutations and its not always clear which ones cause disease. It could even be useful for diagnosing rare conditions and designing new gene therapies. "For all the best evaluations we have, AlphaGenome looks like they pushed [the field] forward a little bit," says David Kelley, a principal investigator at Calico Life Sciences, a company owned by Google's parent company Alphabet. Kelley was not involved with the study but has collaborated with the authors on a previous AI model. "I think the long sequence length that they're able to work with here is definitely one of those major engineering breakthroughs," he says, adding that the new AI is "incremental but real progress." Predicting how a disease manifests from the genome "is an extremely hard problem, and this model is not able to magically predict that," says Žiga Avsec, a research scientist leading DeepMind's genomics initiative. But AlphaGenome can narrow down the pool of possible mutations involved in a disease, making it useful for prioritizing research to pinpoint which gene variants are actually causing problems, he says. DeepMind is not blind to the fact that the model is imperfect. The company's researchers are working to both boost its predictive power and to better report how uncertain those predictions are.
[4]
AI tool AlphaGenome predicts how one typo can change a genetic story
The model can predict changes in 11 biological activities across 1 million DNA letters A new deep-learning AI model may help scientists better decipher the plot of the genetic instruction book and learn how typos alter the story. AlphaGenome, created by Google DeepMind, is the latest in an ever-improving line of AI models built to analyze vast stretches of DNA. The previous front-runner, a model called Borzoi, could predict molecular signposts in stretches of DNA 500,000 bases long. AlphaGenome can analyze 1 million DNA building blocks at a time, researchers report January 28 in Nature. The model may have practical implications for diagnosing rare genetic diseases, identifying cancer-driving mutations, designing synthetic DNA sequences or therapeutic RNAs and better understanding basic biology. "AlphaGenome is not just a bigger model in terms of context length, but it actually is quite a leap forward in its overall utility," says Anshul Kundaje, a computational biologist at Stanford University who develops AI models for genomics. For instance, a genetic change may have no effect on nearby genes but could change activity of genes far away. Because AlphaGenome examines longer stretches of DNA, it is more likely to spot such long-distance relationships. But AlphaGenome isn't perfect. Unpublished data from Kundaje's lab indicates the model struggles with predicting how gene activity changes in individuals. Right now, the model is a tool for uncovering basic biology not something doctors could use to diagnose or treat patients. AlphaGenome has "maxed out" what this type of model can do, Kundaje says. He predicts the next big leap will come from scientists generating new types of data for the model or its descendants to analyze. AlphaGenome can pinpoint biologically important spots down to single base resolution, says Peter Koo, a computational biologist at Cold Spring Harbor Laboratory in New York. That's much higher resolution than Borzoi, which flagged points of biological interest in 32 base-pair bins. That's a big task considering that the model's reference is the 3-billion-base-long human genome, often called a genetic instruction book. The book is actually a multivolume, choose-your-own-adventure, popup encyclopedia. Genes, the short stories of the book, are told in small phrases that can be rearranged, shortened or skipped. In between the story fragments are passages that may contain instructions for how to read a different story entirely. Pages and chapters are intricately folded into each other so that pulling a tab in one passage causes something to pop up chapters away. Much of the book is filled with what many people thought was nonsense but is often essential reading material. Researchers have cataloged a dizzying array of punctuation marks, origami-like creases, syntax swaps, margin scribbles and other types of biological grammar that cells use to make sense of the book. AlphaGenome's task is to take a string of DNA letters and predict how plot points, punctuation and other variations affect 11 distinct biological processes, including RNA splicing, gene activity levels and certain protein-DNA interactions. The model considers 5,930 data points from studies of human DNA and 1,128 in mouse DNA. With those data, the AI can predict how changing a single letter, or base, in the million-base string alters the story. Specialized computational models that predict subsets of these biological functions have been in use for years, but AlphaGenome outperforms them on most measures and does particularly well at identifying some features in different types of cells, the researchers report. For example, AlphaGenome identified gene activity changes in certain cell types 14.7 percent better than Borzoi2. "By doing well on so many different genomic tasks simultaneously, we believe this demonstrates that the model has learned a powerful general representation of DNA sequences and the complex processes these sequences encode," said Natasha Latysheva of Google DeepMind January 27 during a news briefing. The tool could make things easier for researchers who are trying to understand how the genome works, says Judit García González, a human geneticist at the Ichan School of Medicine at Mount Sinai in New York City. Before AlphaGenome, a researcher "might need to use three different tools with their own caveats, and [have] to learn how they work, for predicting say 20 different genomic functional consequences," she says. Now, AlphaGenome unites all those in one tool. AlphaGenome isn't an entirely new invention. It builds on previous models but uses aspects of those models in clever ways. "There is no single innovation in AlphaGenome that one can pinpoint as a critical innovation. It's really a system of lots of tricks and engineering," Koo says. AlphaGenome used one trick called ensemble distillation that Koo's lab has been experimenting with. That strategy pretrains multiple copies of the model each on computationally mutated DNA. Those models serve as teachers to a single student model that averages their outputs. It's like having 60 history professors give their account of an important event, Koo says. "If you consider the consensus across what every historian agrees, what overlaps across their story lines, that is probably what might actually be true." The consensus, he says, "tends to be more reliable than trusting any individual model."
[5]
Google's AlphaGenome wants to do for DNA what AlphaFold did for ...
Google's new deep learning model can predict the effect of small changes to DNA sequences up to one million base pairs in length and is particularly good with non-coding DNA, which has proven especially difficult to understand. The artificial intelligence (AI) tool - called AlphaGenome - offers researchers a way to better understand the human genome and may help scientists develop treatments for disease. AlphaGenome is 'a foundational, high-quality tool that turns the static code of the genome into a decipherable language' Robert Goldstone, Francis Crick Institute Small variations in the human genome can have a big impact on a person's health, causing genetic disorders like cystic fibrosis or certain cancers. Most changes occur in the genome's non-coding regions that make up 98% of the total DNA. These regions influence the expression of genes, rather than coding for proteins, and alterations can often have a range of biological effects, making it hard to predict their impact. AlphaGenome, developed by Google DeepMind, can predict the molecular impact of single base pair variations across whole DNA sequences up to a million base pairs in length. This builds on Google's earlier model, AlphaMissense, which was only able to understand the effects of variations in the coding region of DNA sequences. The new model - trained on human and mouse genome data - takes a DNA sequence as an input and gives predictions on various genetic signals that relate to specific biological functions. This includes gene expression, DNA's accessibility to proteins and where gene splicing occurs. 'The key [benefit] is that you can introduce a mutation to the sequence, changing for example a C [base pair] to a T, and then use the model to compare these differences,' says Google DeepMind researcher Žiga Avsec. What do we mean when we say AI? AlphaGenome matched or outperformed other state-of-the-art models in 25 out of 26 tasks predicting the effects of genetic variations. The team were also able to simulate known DNA mutations responsible for a type of leukaemia, predicting the same results as those observed in the lab. 'Previously, the field required separate models for separate tasks,' says Avsec, adding that earlier models also often had a trade-off between sequence length and resolution. 'AlphaGenome unifies these under one roof.' Natasha Latysheva, a senior research engineer at DeepMind, explains that AlphaGenome may help improve fundamental knowledge about the genome, improve understanding of rare diseases and cancers or help scientists design new DNA sequences to treat specific conditions. AlphaGenome adds to the collection of other AI tools developed by Google DeepMind, which includes the 2024 Nobel prize winning AlphaFold that predicts the 3D shape of proteins. Pushmeet Kohli, who led the work, explains that 'the genome is the recipe and understanding the effect of changing any part of the recipe is what AlphaGenome looks at'. AlphaGenome turns genetic code into 'decipherable language of discovery' Robert Goldstone, head of genomics at the Francis Crick Institute in the UK, believes that AlphaGenome is 'a foundational, high-quality tool that turns the static code of the genome into a decipherable language for discovery', but warns that it 'is not a magic bullet for all biological questions'. Despite the improvements, AlphaGenome still has a number of limitations. Like other models, it struggles to predict the influence of genetic alterations that are more than 100,000 base pairs apart and can only make predictions about DNA sequences from the cell types used to train the model - namely human and mouse. Another issue is interpreting results from the model, explains Jian Zhou, a genomics machine learning researcher at the University of Chicago in the US. 'Even when the model makes accurate predictions, it does not always directly inform us of the underlying biological processes,' he adds. Google DeepMind released a preview of the model for non-commercial research in June last year. Since then, Kohli explains that nearly 3000 scientists in 160 different countries have used AlphaGenome, submitting around 1 million requests each day. He hopes that 'AlphaGenome will continue to be a valuable resource for the scientific community and help scientists better understand genome function and disease biology, and ultimately drive new biological discoveries and ... new treatments'.
[6]
Even Tech Skeptics Can Cheer AI's Promise in Decoding the 'Dark Genome'
The platform's progress is limited by the lack of experimental data for it to train on, and its advancement depends on humans in the lab cataloging critical data to improve the models. Google DeepMind, the artificial intelligence subsidiary of Alphabet, has made another leap in its efforts to illuminate human biology: progress toward using AI to interpret the many still-mysterious chapters in the text of life. DNA sequencing, once a gargantuan feat, is by now cheap and easy. Deciphering the billions of letters in that code, however, is another story -- particularly when it comes to understanding which of the many naturally occurring typos in the text are harmless, and which are implicated in disease. Enter DeepMind's AlphaGenome, a platform that, as is outlined in a Nature paper published this week, seeks to connect those typos to a particular function. This could potentially have lots of real-life applications: speeding up efforts to predict the impact of a rare genetic disease; determining which of the many mutations cropping up in a patient's tumor is driving their cancer; and accelerating the development of genetic medicine, to name a few. It will take a lot more work for those ambitions to be realized. Yet the rapid advances in using AI to imbue meaning in the 3 billion letters in our DNA should still be celebrated. DeepMind has made incredible inroads in using machine learning to translate the text of the genome into biological insights. By far its most prominent advance -- one that in 2024 earned its researchers a Nobel Prize -- has been the development of AlphaFold, a program that predicted the 3-D structure of virtually all known proteins in nature from their genetic sequence. As I've written before, that tremendous feat instantly became a bedrock of drug development. AlphaGenome is tackling a far more complicated problem. Each one of our cells carries the same set of genetic instructions, yet different types -- a heart cell, for example, as opposed to a liver cell -- behave in wildly different ways. This complex orchestration is conducted by the "dark genome," the huge stretches of our DNA that control the genes that determine when, where and by how much various proteins are made. So much of that orchestration remains a mystery -- one with real-world consequences. Every day, oncologists sequence patients' tumors to try to pinpoint the drivers of their cancer, tailor treatment, and predict the course their disease. Yet doctors "get information we don't know what to do with all the time," says Omar Abdel-Wahab, a physician-researcher at Memorial Sloan Kettering Cancer Center. When they spot a new typo in someone's DNA, they want to know if its function is important or not. That's where AlphaGenome comes in. It can predict nearly a dozen types of tasks from a sequence, such as whether it tunes the volume on a gene or where a gene is snipped apart. Some of those functions are already addressed by existing tools used by researchers, and in the Nature paper, DeepMind scientists showed that AlphaGenome performed as well as or better than all of them. (Abdel-Wahab, for example, is already using a tool called Splice AI to predict whether a patient's mutations are relevant, and told me he is impressed that AlphaGenome appears to outperform it.) The work comes with plenty of caveats. For starters, DeepMind's platform works well for predicting some gene functions, but not all of them. Scientists tell me that for now, it might best be considered a filter rather than a finder -- that is, it can efficiently narrow down the possible disease drivers, rather than confidently pinpoint the culprit. Sign up for the Bloomberg Opinion bundle Sign up for the Bloomberg Opinion bundle Sign up for the Bloomberg Opinion bundle Get Matt Levine's Money Stuff, John Authers' Points of Return and Jessica Karl's Opinion Today. Get Matt Levine's Money Stuff, John Authers' Points of Return and Jessica Karl's Opinion Today. Get Matt Levine's Money Stuff, John Authers' Points of Return and Jessica Karl's Opinion Today. Bloomberg may send me offers and promotions. Plus Signed UpPlus Sign UpPlus Sign Up By submitting my information, I agree to the Privacy Policy and Terms of Service. And right now, AlphaGenome can only make predictions about certain types of cells, a limitation that has less to do with the power of the algorithm than the lack of experimental data for it to train on. That's a problem that can't be solved by ingenious engineering alone, says Peter Koo, a professor at Cold Spring Harbor Laboratory who develops deep learning methods for connecting genes to their function. "They're pushing us towards the plateau of what we can achieve with existing data," Koo says. Progress, ironically, depends on humans in the lab -- biologists who can catalog the most critical data AlphaGenome needs to advance. That work should be done thoughtfully, with an eye toward prioritizing experiments that will help improve the models, Koo says. As the scientific community learns about where the DeepMind tool can be most useful and builds out the data needed to make it even better, Although DeepMind has made the tool freely available for non-commercial use, it's easy to imagine those lines blurring as academic labs make discoveries based in part on its use -- even as their own data might have contributed to improving its accuracy. Much like AlphaFold, AlphaGenome would not be possible without access to large, publicly available, publicly funded datasets. At a moment when funding for government-sponsored research is tenuous, the advance should be a reminder of the value in the bread-and-butter work performed by scientists in the US. The impact can stretch far beyond one project or one patient -- it could one day be the foundation for the next game-changing technology. More From Bloomberg Opinion: Want more Bloomberg Opinion? OPIN <GO>. Or subscribe to our daily newsletter.
[7]
AI model from Google DeepMind reads recipe for life in our DNA
An AI model developed by Google's DeepMind could transform our understanding of DNA - the complete recipe for building and running the human body - and its impact on disease and medicine discovery, according to researchers. Called AlphaGenome, the model could help scientists discover why subtle differences in our DNA put us at risk of conditions such as high blood pressure, dementia and obesity. It could also dramatically accelerate our understanding of genetic diseases and cancer. The developers of the model acknowledge it's not perfect, but experts have described it as "an incredible feat" and "a major milestone". "We see AlphaGenome as a tool for understanding what the functional elements in the genome do, which we hope will accelerate our fundamental understanding of the code of life," says Natasha Latysheva, research engineer at DeepMind. The human genome is made up of three billion letters of DNA code - represented by the letters A,C,G and T. Around 2% of it are genes which code for all the proteins the body needs to grow and function. The remaining 98%, which is less well understood, is labelled the 'dark genome'. It plays a crucial role in organising how genes are used in the body and is where many mutations linked to disease are found. AlphaGenome can analyse one million letters of code at a time, helping to unravel the 'dark genome'. It can predict where the genes are, but also what the 'dark genome' is influencing. For example, how it affects gene expression (whether a gene is highly active or being suppressed) and gene splicing (the tool the body uses to make different proteins from a single gene). Crucially, the model can predict the impact of changing even a single letter in genetic code. Latysheva said she was "really excited" by the AI model's potential to understand which mutations cause disease and help pinpoint the cause of rare genetic diseases. The AI model could be used to "add another piece of the puzzle for the discovery of drug targets and ultimately the development of new drugs", she added. Ultimately, it could also be used in synthetic biology and the design of new sequences of DNA which could be used in gene therapies. AlphaGenome has been described in the journal Nature, but was made available for non-commercial use last year and 3,000 scientists have since used the tool. Dr Gareth Hawkes, from the University of Exeter, is using it to explore how mutations could be altering our risk of obesity and diabetes. Studies that sequenced the entire genetic code of tens of thousands of people have identified variants linked to the conditions, but they are often in the dark genome. "They're directly impacting some important piece of biology that we don't really understand," Hawkes told the BBC. Using AlphaGenome allows researchers to rapidly predict what those variants are up to so they can be tested in the lab. Hawkes said: "Those predictions will help to inform which biological processes those genetic variants might be impacting, and potentially lead to drug developments. "I wouldn't say the dark side of the genome is solved by AlphaGenome, but it's a big leap. I'm really excited." Cancer is another field where the AI model could accelerate research. AlphaGenome has been used to predict which mutations are fuelling cancer and are also the potential targets of treatment, and which mutations are incidental. Dr Robert Goldstone, head of genomics at the Francis Crick Institute, said the model was a "major milestone in the field of genomic AI" and the breakthrough was "an incredible technical feat" for its "ability to predict gene expression from DNA sequence alone". Prof Ben Lehner, the head of generative and synthetic genomics at the Wellcome Sanger Institute, said they had tested AlphaGenome in more than half a million experiments and it was performing very well. But he said it was "far from perfect" and there was still a lot of work to do. "It's a really exciting time with three areas where the UK is world-leading - genomics, biomedical research and AI - combining to transform biology and medicine," Prof Lehner said. The team at DeepMind won the Nobel Prize for Chemistry in 2024 for their work on AlphaFold - an AI system that predicts the 3D structure of proteins in the body. "I think we are at the start of a new era of scientific progress, and AI is going to enable a number of different breakthroughs," says Pushmeet Kohli, vice president of science and strategic initiatives at Google DeepMind. AlphaGenome doesn't work like large language models (such as ChatGPT) that predict the next word in a sequence. Instead, it is a "sequence-to-function model" looking at how changes in the text affect the meaning at the end. It was trained on publicly available databases of human and mouse cell experiments. There is general agreement that the AI model needs refining. It is less accurate in some areas such as predicting how genes are regulated over long distances (more than 100,000 letters of code away). The team also want to improve the accuracy of the model in different tissues. A neuron in the brain, for example, has the same genetic code as a beating heart cell, but each has different properties based on the way the genetic instructions are being used in each cell type.
[8]
DeepMind's New AI Can Read a Million DNA Letters at Once -- and Actually Understand Them
AlphaGenome is reportedly the most comprehensive and accurate DNA sequence model developed to date. Artificial intelligence has gotten a bad reputation lately, and often for good reason. But a team of scientists at Google's DeepMind now claims to have found a revolutionary use case for AI: helping humanity unravel the "dark matter" of our genome more effectively than ever before. In a study published today in Nature, DeepMind researchers debuted their deep learning model, dubbed AlphaGenome. Compared to existing models, AlphaGenome can predict the function of much longer sequences of DNA while still maintaining a similar level of accuracy, the researchers claim. The team is hopeful its model can become a valuable tool to analyze how subtle variations in human DNA can affect our health and biology, particularly in the vast majority of the genome that works silently in the background. A guide to our genetic dark matter Our DNA contains the instructions for building and regulating every biological aspect of ourselves. But only a tiny portion of our genes, 2% or so, actually carry the code for the tens to hundreds of thousands of proteins that perform the functions a body needs to survive, such as insulin or collagen. The other 98% of our DNA is made of non-coding regions, more eloquently known as the dark matter of our genome. Scientists once assumed our genetic dark matter was comprised of worthless junk DNA, but we now know that it contains sequences vital to regulating our protein-making genes. While scientists have mapped out most of the human genome, we still know very little about how many of these genes work, especially those found in non-coding regions; we're also largely in the dark about how variations in these genes can affect their functioning. Long before AI became a cultural buzzword (and punching bag), scientists had been using deep learning models -- trained on lab data -- to more efficiently sift through the mountains of the human genome and to predict a gene or DNA sequence's function. But DeepMind researchers say AlphaGenome is the most comprehensive and accurate DNA sequence model to date. The DeepMind researchers trained the model on both human and mouse genomes. It can reportedly analyze up to 1 megabase (Mb) -- about 1 million DNA letters -- at a time, compared to older models capable of analyzing upwards of 500 kilobases (kb), though at some cost. From that sequence, the model is said to "predict thousands of functional genomic tracks." These tracks don't just include how a gene or DNA sequence is expressed but also other less visible functions. These include the interactions between coding and non-coding regions of DNA, or the structure of chromatins (the loose packages of genetic material typically found in a cell; chromosomes are the more neatly packaged version). In the paper, the researchers also detailed how AlphaGenome matched or outperformed other existing AI models in 25 out of 26 tests measuring how well it could predict the effects of a genetic variant. More than just accuracy, however, the model can also do more at once; it can simultaneously predict nearly 6,000 human genetic signals tied to specific functions, according to the researchers. The future of AI genomics At least some outside scientists have praised the capabilities of AlphaGenome, while noting that it can't solve every lingering mystery about our genetic code just yet. "At the Wellcome Sanger Institute we have tested AlphaGenome using over half a million new experiments and it does indeed perform very well," Ben Lehner, head of Generative and Synthetic Genomics at the University of Cambridge's Wellcome Sanger Institute, told the Science Media Center. "However, AlphaGenome is far from perfect and there is still a lot of work to do. AI models are only as good as the data used to train them. Most existing data in biology is not very suitable for AI -- the datasets are too small and not well standardized." All that said, the DeepMind researchers -- and others in the field -- believe AlphaGenome marks a true milestone in AI genomics, one that could help make the technology practical for broader use. They argue that AlphaGenome, or similar models, could now be used to better diagnose rare genetic diseases, identify mutations that drive cancer, or uncover new drug targets.
[9]
Google DeepMind launches AI tool to help identify genetic drivers of disease
AlphaGenome can analyse up to 1m letters of DNA code at once and could pave way for new treatments Researchers at Google DeepMind have unveiled their latest artificial intelligence tool and claimed it will help scientists identify the genetic drivers of disease and ultimately pave the way for new treatments. AlphaGenome predicts how mutations interfere with the way genes are controlled, changing when they are switched on, in which cells of the body, and whether their biological volume controls are set to high or low. Most common diseases that run in families, including heart disease and autoimmune disorders, as well as mental health problems, have been linked to mutations that affect gene regulation, as have many cancers, but identifying which genetic glitches are to blame is far from straightforward. "We see AlphaGenome as a tool for understanding what the functional elements in the genome do, which we hope will accelerate our fundamental understanding of the code of life," Natasha Latysheva, a DeepMind researcher, told a press briefing on the work. The human genome runs to 3bn pairs of letters - the Gs, Ts, Cs and As that comprise the DNA code. About 2% of the genome tells cells how to make proteins, the building blocks of life. The rest orchestrates gene activity, carrying the crucial instructions that dictate where, when and how much individual genes are switched on. The researchers trained AlphaGenome on public databases of human and mouse genetics, enabling it to learn connections between mutations in specific tissues and their impact on gene regulation. The AI can analyse up to 1m letters of DNA code at once and predict how mutations will affect different biological processes. The DeepMind team believes the tool will help scientists map out which strands of genetic code are most essential for the development of particular tissues, such as nerve and liver cells, and pinpoint the most important mutations for driving cancer and other diseases. It could also underpin new gene therapies by allowing researchers to design entirely new DNA sequences - for example, to switch on a certain gene in nerve cells but not in muscle cells. Carl de Boer, a researcher at the University of British Columbia in Canada, who was not involved in the work, said: "AlphaGenome can identify whether mutations affect genome regulation, which genes are impacted and how, and in what cell types. A drug could then be developed to counteract this effect. "Ultimately, our goal is to have models that are so good we don't have to do an experiment to confirm their predictions. While AlphaGenome represents a significant innovation, achieving this goal will require continued work from the scientific community." Some scientists have already begun using AlphaGenome. Marc Mansour, a clinical professor of paediatric haemato-oncology at UCL, said it marked a "step change" in his work to find genetic drivers for cancer. Gareth Hawkes, a statistical geneticist at the University of Exeter, said: "The non-coding genome is 98% of our 3bn base pair genome. We understand the 2% fairly well, but the fact that we've got AlphaGenome that can make predictions of what this other 2.94bn base pair region is doing is a big step forward for us."
[10]
Google's new AI tool decodes DNA mutations. Here's how it works
A new AI model by Google DeepMind can decipher DNA and predict mutations, opening new doors for disease research. Our DNA is made of millions of combinations of the genomes that create the human body. Even the smallest changes in these sequences, or in how they act, can change the functioning of the whole body and cause diseases such as cancer. AlphaGenome, Google's new artificial intelligence (AI) tool, can read large stretches of DNA and predict how different sections behave and how changes could lead to diseases. Using deep learning and inspired by how the brain processes information, it is designed to help scientists understand how DNA works. Google's new tool can help decode how DNA controls genes by predicting what long stretches of genetic code do. "We believe AlphaGenome can be a valuable resource for the scientific community, helping scientists better understand genome function, disease biology, and ultimately, drive new biological discoveries and the development of new treatments," Google DeepMind said. The model reads up to one million DNA letters with single-letter precision - something impossible with previous tools. DNA consists of long chains made from four basic chemical building blocks called nucleotides, each identified by a letter: A, C, G, and T. It works as an instruction manual for making and controlling every cell. Only about two percent of human DNA directly codes for proteins, the building blocks that do most of the work in our cells. The remaining 98 percent has long been dismissed as "junk DNA"; however, far from being useless, these sequences act like control panels regulating how the other two percent works. They guide when, where, and how much genes turn on or off, respond to environmental signals, and influence RNA splicing, a system that joins sequences of letters and allows the same gene to produce different readings. Many disease-linked variants hide here, affecting gene activity without altering proteins. AlphaGenome is the first deep learning model able to target this part of DNA and predict its functioning. The model can estimate how small genetic changes, called variants, can affect gene activity or interrupt normal processes linked to diseases such as cancer. As a real-life example, the researchers focused on a type of acute leukaemia, a cancer of the white blood cells, where immature T-cells, immune fighters, grow out of control. Some leukaemia cases are caused by small changes in DNA that don't change a protein itself, but instead change how strongly or when certain genes turn on. The AlphaGenome model compared the normal DNA sequence with the mutated one, and predicted how likely the mutation is to increase the activity of nearby genes. The model is currently available for scientists for free for non-commercial research; it is a research tool, not meant to be clinically used. The research team sees multiple uses for the new model. In molecular biology, it can work like a virtual lab tool, allowing scientists to test ideas by simulation before doing expensive experiments. In biotechnology, it can help design genetic therapies or improve molecules that target specific tissues. "DeepMind's AlphaGenome represents a major milestone in the field of genomic AI," said Robert Goldstone, head of genomics at the Francis Crick Institute. He added that the level of resolution that the new model allows is a breakthrough that moves the technology from theoretical interest to practical utility, allowing scientists to programmatically study and simulate the genetic roots of complex disease. "AlphaGenome is not a magic bullet for all biological questions, but it is a foundational, high-quality tool that turns the static code of the genome into a decipherable language for discovery," Goldstone added. However, scientists warn that, like all AI models, AlphaGenome is only as good as the data used to train it. "Most existing data in biology is not very suitable for AI - the datasets are too small and not well standardised", said Ben Lehner, head of generative and synthetic genomics at the Wellcome Sanger Institute in the United Kingdom. According to him, the most important challenge right now is how to generate the data to train the next generation of AI models.
[11]
Google unveils AI tool probing mysteries of human genome
Paris (France) (AFP) - Google unveiled an artificial intelligence tool Wednesday that its scientists said would help unravel the mysteries of the human genome -- and could one day lead to new treatments for diseases. The deep learning model AlphaGenome was hailed by outside researchers as a "breakthrough" that would let scientists study and even simulate the roots of difficult-to-treat genetic diseases. While the first complete map of the human genome in 2003 "gave us the book of life, reading it remained a challenge", Pushmeet Kohli, vice president of research at Google DeepMind, told journalists. "We have the text," he said, which is a sequence of three billion nucleotide pairs represented by the letters A, T, C and G that make up DNA. However "understanding the grammar of this genome -- what is encoded in our DNA and how it governs life -- is the next critical frontier for research," said Kohli, co-author of a new study in the journal Nature. Only around two percent of our DNA contains instructions for making proteins, which are the molecules that build and run the body. The other 98 percent was long dismissed as "junk DNA" as scientists struggled to understand what it was for. However this "non-coding DNA" is now believed to act like a conductor, directing how genetic information works in each of our cells. These sequences also contain many variants that have been associated with diseases. It is these sequences that AlphaGenome is aiming to understand. A million letters The project is just one part of Google's AI-powered scientific work, which also includes AlphaFold, the winner of 2024's chemistry Nobel. AlphaGenome's model was trained on data from public projects that measured non-coding DNA across hundreds of different cell and tissue types in humans and mice. The tool is able to analyse long DNA sequences then predict how each nucleotide pair will influence different biological processes within the cell. This includes whether genes start and stop and how much RNA -- molecules which transmit genetic instructions inside cells -- is produced. Other models already exist that have a similar aim. However they have to compromise, either by analysing far shorter DNA sequences or decreasing how detailed their predictions are, known as resolution. DeepMind scientist and lead study author Ziga Avsec said that long sequences -- up to a million DNA letters long -- were "required to understand the full regulatory environment of a single gene". And the high resolution of the model allows scientists to study the impact of genetic variants by comparing the differences between mutated and non-mutated sequences. "AlphaGenome can accelerate our understanding of the genome by helping to map where the functional elements are and what their roles are on a molecular level," study co-author Natasha Latysheva said. The model has already been tested by 3,000 scientists across 160 countries and is open for anyone to use for non-commercial reasons, Google said. "We hope researchers will extend it with more data," Kohli added. 'Breakthrough' Ben Lehner, a researcher at Cambridge University who was not involved in developing AlphaGenome but did test it, said the model "does indeed perform very well". "Identifying the precise differences in our genomes that make us more or less likely to develop thousands of diseases is a key step towards developing better therapeutics," he explained. However AlphaGenome "is far from perfect and there is still a lot of work to do", he added. "AI models are only as good as the data used to train them" and the existing data is not very suitable, he said. Robert Goldstone, head of genomics at the UK's Francis Crick Institute, cautioned that AlphaGenome was "not a magic bullet for all biological questions". This was partly because "gene expression is influenced by complex environmental factors that the model cannot see", he said. However the tool still represented a "breakthrough" that would allow scientists to "study and simulate the genetic roots of complex disease", Goldstone added.
[12]
Google DeepMind open-sources AlphaGenome medical research model
Google DeepMind open-sources AlphaGenome medical research model Google DeepMind today open-sourced AlphaGenome, an artificial intelligence model that researchers can use to study biological processes. The Alphabet Inc. unit first debuted the algorithm in June. Until now, it was only accessible through an application programming interface limited to non-commercial research use cases. According to DeepMind the API has been adopted by more than 3,000 scientists and processes about 1 million requests per day. The Alphabet unit created AlphaGenome to accelerate DNA-focused medical research projects. According to the company, the model can help scientists better understand the role of DNA in biological processes and study diseases. DNA contains instructions that cells use to produce proteins. Proteins, the basic building blocks of life, facilitate interactions between cells and power numerous other biological processes. Researchers can use AlphaGenome to understand how changes to protein production instructions impact health. The model also lends itself to studying certain related phenomena. DNA contains numerous protein production instructions, but cells use only a tiny percentage of them on a day-to-day basis. AlphaGenome makes it easier to determine which instructions are used by a cell in a given scenario. A DNA molecule is made up of segments called base pairs that are arranged in a double helix. Those base pairs are the instructions that cells use to make proteins. Each base pair comprises two nitrogen-based chemical compounds, or nucleobases, that are linked together by hydrogen atoms. According to DeepMind, AlphaGenome can map out the molecular properties of DNA sequences with up to 1 million base pairs. That's significantly more than the context window of earlier models. AlphaGenome also generates higher-resolution molecular property predictions, which provides researchers with more accurate medical data. According to DeepMind, the model comprises three modules based on different AI architectures. Each one performs a different set of molecular calculations. The first module is a convolutional neural network, a type of AI that is mainly used for image and video analysis tasks. It's responsible for carrying out the initial set of tasks involved in analyzing base pairs. From there, the processing results are refined by transformers. A third set of artificial neurons turns the data into molecular property predictions that scientists can use in their research. In a Nature paper released today, DeepMind detailed that AlphaGenome outperformed competing models across 25 of the 26 evaluations it ran internally. Furthermore, the model can provide that performance using a relatively limited amount of hardware. Researchers can run AlphaGenome on only a single H100 graphics processing unit. The model's release comes five years after DeepMind introduced its seminal AlphaFold neural network. The latter algorithm can automatically predict a protein's shape, a task that historically took months of manual work. AlphaFold's co-creators won one half of the 2025 Nobel Prize in chemistry.
[13]
DeepMind's AlphaGenome: How AI Is Reading DNA's Code of Life
Scientists have traditionally referred to DNA as the blueprint to life. Another potent new artificial intelligence (AI) application, the AlphaGenome created by DeepMind, a division of Google, is enabling researchers to perceive that recipe in a brand-new approach. This model is published in the scientific journal Nature and can be used in research where human DNA sequences can be analyzed, and thus, it is able to predict how the sequence regulates biological functioning. The entire genome of our cells, the human genome, contains more than three billion chemical letters in the form of A, C, G, and T. Even those letters directly coded to proteins, the molecular machines that perform the fundamental functions of life, are only about 2%. The other 98 percent, commonly referred to as the dark genome, does not produce proteins but holds instructions on the timing and location of genes being switched on or off. These health- and disease-critical regulatory regions are, however, much more difficult to interpret. To address this problem, AlphaGenome was developed. Conventional genetics models were only capable of analyzing small fragments of DNA or a particular kind of biological signal. The AlphaGenome is different: it has the ability to accept up to a million letters of DNA simultaneously and predict a large variety of biological activities associated with control of the genes in one run. AlphaGenome can provide high-resolution predictions regarding a variety of aspects of genome function once it has been taught. These include the beginning and ending locations of genes, the amount of RNA produced by a gene (a measure of its activity), the splicing of RNA molecules, and the accessibility of various DNA sections to proteins. Importantly, the model can also estimate how a single-letter alteration in DNA, referred to as a genetic variant, would affect biological processes. This is significant since most genetic variants associated with diseases are not found in genes that encode proteins but instead are found in the regulatory regions. In the past, researchers were forced to use time and costly experiments in the laboratory to evaluate the impacts of each variant. Using AlphaGenome, they are able to work out which variants should be given priority and to focus experimental effort on where it will be most educative. Researchers from all across the world are already using the tool. More than 3,000 scientists have used DeepMind's API to access AlphaGenome in order to find answers to questions about how DNA alterations can impact disease risk, drug response, and biological processes in general. Some teams are investigating the model in relation to uncommon genetic illnesses, diabetes, and cancer. AlphaGenome is not a medical diagnosis; it does not diagnose disease on its own or forecast the health of an individual-- regardless of how promising it may be. It is, in fact, a research tool that has accelerated discovery and assisted scientists in figuring out the complicated language of the genome. Its predictions are yet to be put to the test in the laboratory. AlphaGenome is the continuation of DeepMind working with AI in the biological field. The previous system, AlphaFold, of the company had transformed the protein structure prediction and won a Nobel Prize in Chemistry due to its contribution to the scientific community. AlphaGenome also makes a similarly ambitious move in its effort to solve the regulating of the genome, one of the most challenging puzzles in biology. In the long run, tools like AlphaGenome could help researchers uncover new therapeutic targets, improve our understanding of how genetic variation leads to disease, and even aid in designing synthetic DNA for future gene therapies. For now, it stands as a milestone in the use of AI to read and interpret the biological code that makes each of us who we are.
[14]
AlphaGenome explained: How Google DeepMind is using AI to rewrite genomics research
The history of genomics has long been a story of reading a book where only every fiftieth page made sense. While the Human Genome Project gave us the full sequence of human DNA over two decades ago, we were largely illiterate in the language of the "non-coding" genome - the 98% of our genetic code that does not build proteins but dictates when, where, and how those proteins are made. This vast expanse, often dismissed as "dark matter," is where the instructions for human life and the triggers for complex diseases are hidden. On January 28, 2026, Google DeepMind published a landmark paper in Nature detailing AlphaGenome, an AI model that finally provides a high-resolution map for this genetic wilderness. Also read: OpenAI plans social network: Can Sam Altman stop bots from entering? Bridging the gap between context and detail For years, computational biology was trapped in a frustrating trade-off. AI models could either focus on a tiny fragment of DNA with intense scrutiny or look at a broad landscape with blurry vision. This was a significant hurdle because human biology is inherently "long-range"; a mutation in a regulatory switch at one end of a chromosome can trigger a gene located millions of base pairs away. AlphaGenome shatters this limitation by utilizing a hybrid architecture that combines the spatial efficiency of U-Nets with the long-range "attention" of Transformers. The result is a model capable of processing sequences up to one million base pairs long while maintaining the ability to pinpoint the biological consequence of a single-letter change. By "reading" DNA in such massive windows, AlphaGenome can observe the complex folding and interactions that govern gene expression in a way that previous models, limited to much shorter sequences, simply could not. In internal benchmarks, this allowed AlphaGenome to outperform existing state-of-the-art models in nearly every category, particularly in predicting how specific genetic variants affect gene expression. A multimodal engine for precision medicine AlphaGenome does not just look for patterns; it simulates the fundamental mechanics of life. It is trained to simultaneously predict over 7,000 distinct genetic signals across both humans and mice. This includes the incredibly complex process of RNA splicing, where cellular machinery cuts and pastes genetic instructions. Errors in this process are the root cause of devastating conditions like spinal muscular atrophy and cystic fibrosis. By explicitly modeling these "splice junctions," AlphaGenome allows researchers to see exactly how a mutation will disrupt the final protein product before a patient ever sets foot in a lab. Also read: Clawdbot security issues: Private messages to AI agent can leak This predictive power turns the model into a "virtual laboratory." Traditionally, identifying whether a rare genetic variant is harmful or benign requires years of expensive "wet lab" experimentation. AlphaGenome can evaluate the impact of these "variants of uncertain significance" in less than a second on a modern GPU. For a clinician trying to diagnose a child with a rare, undiagnosed condition, this tool moves the needle from guesswork to actionable data, identifying the specific "broken switch" responsible for the disease. Open-sourcing the genetic revolution Perhaps the most significant aspect of AlphaGenome is its availability. Following the precedent set by AlphaFold, DeepMind has open-sourced the model and its code for non-commercial research. This move democratizes the front lines of genetic science. It ensures that a researcher at a university in India or a non-profit lab in Brazil has the same analytical power as a multi-billion dollar pharmaceutical company. As we move into an era of "Agentic AI," where models act as autonomous collaborators, AlphaGenome represents the foundational layer for the next decade of discovery. It is already being used to design synthetic "enhancers" - custom DNA sequences that can turn on a therapeutic gene in a specific organ without affecting the rest of the body. By providing the dictionary for the dark genome, AlphaGenome is not just helping us read our DNA; it is teaching us how to write a healthier future.
Share
Share
Copy Link
Google DeepMind unveiled AlphaGenome, a deep-learning AI model that predicts how mutations in non-coding DNA affect gene expression across sequences up to one million base pairs long. The tool outperforms existing models in 25 out of 26 tasks and is already being used by nearly 3,000 scientists worldwide to investigate rare diseases, cancer mutations, and design new gene therapies.
AlphaGenome Addresses Biology's Grand Challenge
Google DeepMind has released AlphaGenome, a deep-learning AI model designed to decode the 98% of the human genome that doesn't produce proteins
1
. Described in Nature on January 28, this research tool can analyze DNA sequences up to one million base pairs in length and predict how mutations in non-coding DNA affect gene expression and other biological processes4
. The model represents what researchers call a "Swiss Army knife for exploring non-coding DNA," offering unprecedented precision in modeling intricate genomic processes2
.
Source: Digit
Deciphering non-coding regions has long been one of biology's most stubborn challenges. Once dismissed as "junk DNA," these sequences are now understood to be crucial for determining when, where, and how genes are turned on and off
3
. Mutations in these areas are especially vexing to researchers seeking to uncover the genetic basis for rare, often fatal genetic diseases. "These are variants that, to be quite honest, often get triaged," says Eric Klee, a bioinformatician at the Mayo Clinic who tested AlphaGenome at the Undiagnosed Hackathon in September 20251
.How AlphaGenome Predicts Genetic Changes Across Multiple Tasks
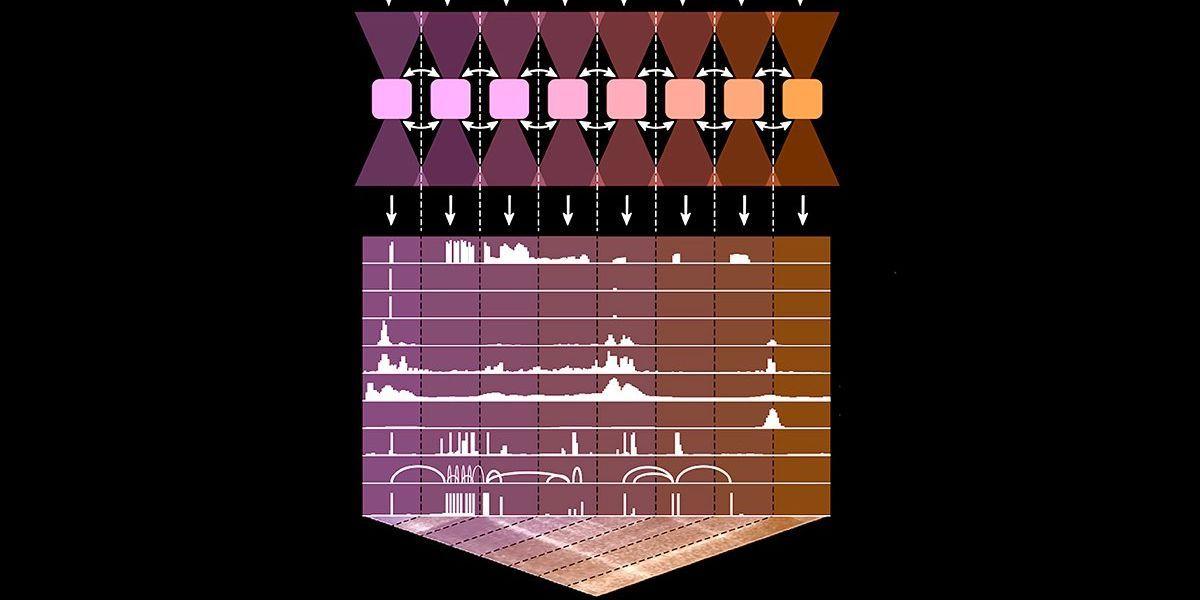
AlphaGenome takes a DNA sequence as input and predicts 11 types of biological signals that help determine how genes function inside cells
2
. These include whether a gene is activated or silenced, where gene activity begins, how genetic messages are edited, how tightly DNA is packed, which regulatory proteins bind to it, and how distant regions of the genome interact. The model can pinpoint biologically important spots down to single base resolution, a significant improvement over its predecessor Borzoi, which identified points of biological interest in 32 base-pair bins4
.
Source: ET
The tool matched or outperformed other state-of-the-art models in 25 out of 26 tasks predicting the effects of genetic variations
5
. For example, AlphaGenome identified gene activity changes in certain cell types 14.7 percent better than Borzoi24
. The team also successfully simulated known DNA mutations responsible for a type of leukemia, predicting the same results observed in laboratory experiments5
.From AlphaFold to AlphaGenome: Building a Molecular Prediction Platform
AlphaGenome extends Google DeepMind's growing stable of biological models, which includes the Nobel Prize-winning AlphaFold for protein structure prediction, AlphaMissense for analyzing mutations in protein-coding regions, and AlphaProteo for designing proteins that bind to specific molecular targets
2
. "The genome is the recipe and understanding the effect of changing any part of the recipe is what AlphaGenome looks at," explains Pushmeet Kohli, vice president of science at Google DeepMind5
.
Source: IEEE
While AlphaMissense focused on the small fraction of the genome that codes for proteins, AlphaGenome tackles the far larger regulatory landscape
3
. This vertical integration across genomics creates a comprehensive platform for molecular prediction that could unlock new diagnostic capabilities and therapeutic strategies. "All these different models are solving key problems that are relevant for understanding biology," Kohli notes2
.Related Stories
Real-World Applications in Rare Disease Diagnosis
Nearly 3,000 scientists in 160 countries have already used AlphaGenome since Google DeepMind released a preview for non-commercial research in June last year, submitting around one million requests daily
5
. At the Undiagnosed Hackathon, where more than 100 researchers tackled 29 undiagnosed conditions, AlphaGenome was deployed to investigate several cases1
. The event, organized by the Wilhelm Foundation—a charity founded by parents who lost three of their four children to an undiagnosed disease—aims to help the approximately 350 million people worldwide living with undiagnosed rare conditions.Klee tested AlphaGenome's predictions on a variant his team had linked to an individual's diagnosis before the hackathon. Experimental work showed the mutation altered gene expression in cardiac cells but not neural cells, aligning with the patient's symptoms. AlphaGenome's predictions supported this conclusion
1
. While none of the AlphaGenome predictions at the September 2025 hackathon led directly to a diagnosis, six of the 29 conditions were diagnosed using other approaches, demonstrating how AI tools complement traditional methods.Limitations and Future Directions in Genomics Research
Despite its advances, AlphaGenome faces notable constraints. The model's training data draw largely from bulk tissue datasets, limiting reliability in rare cell types or specific developmental stages. "Generalization to new cell types is a huge limitation," notes Christina Leslie, a computational biologist at Memorial Sloan Kettering Cancer Center
2
. The tool also struggles to capture distant effects when regulatory regions are hundreds of thousands to millions of DNA letters away from their target genes.Additionally, while the model makes accurate predictions, it doesn't always directly inform researchers of the underlying biological processes, explains Jian Zhou, a genomics machine learning researcher at the University of Chicago
5
. Robert Goldstone, head of genomics at the Francis Crick Institute, describes AlphaGenome as "a foundational, high-quality tool that turns the static code of the genome into a decipherable language for discovery," but cautions it "is not a magic bullet for all biological questions"5
. Researchers suggest the next leap will come from generating new types of data for the model to analyze, as AlphaGenome has "maxed out" what this type of architecture can achieve4
.References
Summarized by
Navi
[3]
[5]
Related Stories
DeepMind's AlphaGenome: Decoding the 'Dark Matter' of DNA with AI
26 Jun 2025•Science and Research

AI Model Predicts Gene Activity in Human Cells, Transforming Biological Research
09 Jan 2025•Science and Research

Evo-2: AI Model Revolutionizes Genomic Research and DNA Generation
20 Feb 2025•Science and Research

Recent Highlights
1
OpenAI Releases GPT-5.4, New AI Model Built for Agents and Professional Work
Technology

2
Pentagon's Anthropic showdown exposes who controls AI guardrails in military contracts
Policy and Regulation

3
Anthropic challenges Pentagon supply chain risk label in court over AI usage restrictions
Policy and Regulation





