AI Model Revolutionizes Drug Safety by Predicting Human Toxicity Before Clinical Trials
2 Sources
2 Sources
[1]
Machine learning advances human-centric drug toxicity prediction
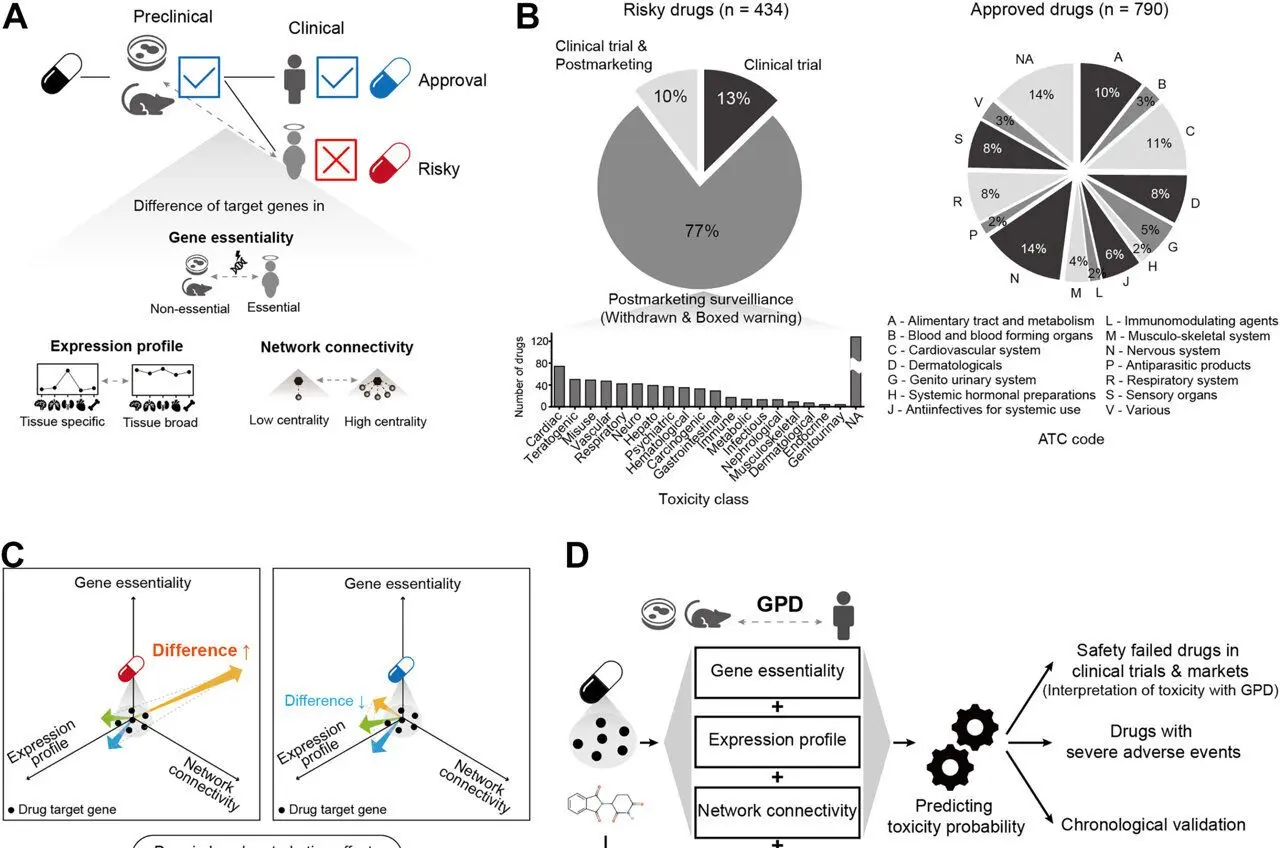
Pohang University of Science & Technology (POSTECH)Nov 7 2025 In the UK, there was a case where TGN1412, an immunotherapy under development, triggered a cytokine storm within hours of administration to humans, leading to multiple organ failure. Another example, Aptiganel, a stroke drug candidate, was also highly effective in animals but was discontinued in humans due to side effects such as hallucinations and sedation. Even though drugs considered safe in preclinical tests can be fatal in human clinical trials. A machine-learning-based technology has been developed to learn these differences and preemptively identify potentially dangerous drugs before clinical trials. A research team led by Professor Sanguk Kim of the Department of Life Sciences and the Graduate School of Artificial Intelligence at POSTECH, along with Dr. Minhyuk Park and Mr. Woomin Song of the Department of Life Sciences, and Mr. Hyunsoo Ahn of the Graduate School of Artificial Intelligence, has developed a technology that uses machine learning to predict drug side effects in humans. This study was recently published online in the international medical journal eBioMedicine. During the development of new drugs, those that pass preclinical trials often show unexpected toxicity in humans. This issue arises from differences in biological responses between humans and animals. For example, chocolate is generally safe for humans but toxic to dogs. Similarly, a drug that is safe in mice does not necessarily mean it is safe for humans. To date, this "cross-species difference" has been a major reason for failures in new drug development. The research team focused on the "Genotype-Phenotype Difference (GPD)," the biological differences between cells, mice, and humans. They analyzed how genes targeted by drugs function differently in humans and preclinical models, focusing on three key factors: first, the gene's perturbation impact on survival (essentiality); second, the pattern of gene expression in different tissues; and third, the connectivity of genes within biological networks. Validation using data from 434 hazardous drugs and 790 approved drugs revealed that GPD characteristics were significantly associated with drug failure due to toxicity in humans. Predictive power was significantly improved over relying on drug chemical data, with the area under the curve (AUPRC1) increasing from 0.35 to 0.63, and the area under the curve (AUROC2) increasing from 0.50 to 0.75. The developed AI model demonstrated superior predictive performance compared to existing state-of-the-art models. Furthermore, it demonstrated practicality in "chronological validation," which alerts users to drugs facing market withdrawal due to toxicity. After training the prediction model on only drug data up to 1991, it correctly predicted drugs expected to be withdrawn from the market after 1991, achieving 95% accuracy. The significance of this study is that it bridges the "translation gap" between preclinical and clinical trials by quantifying biological differences in cells, preclinical animal models, and humans. Pharmaceutical companies can reduce development costs and time by screening out high-risk candidates before clinical trials, while also improving patient safety. The model's effectiveness is expected to increase as more relevant data and annotations accumulate. This is the first attempt to incorporate differences in genotype-phenotype relationships for drug toxicity prediction. Our framework enables early identification of high-risk drugs in clinical development. This approach holds promise for reducing development costs, improving patient safety, and increasing the success rate of therapeutic approvals. Co-first authors Dr. Min-hyuk Park and Mr. Woomin Song stated, "The human-centered toxicity prediction model will be a very practical tool in new drug development. We anticipate that pharmaceutical companies will be able to screen out high-risk drugs in advance at the preclinical stage, thereby improving development efficiency." Professor Sanguk Kim, Department of Life Sciences and the Graduate School of Artificial Intelligence at POSTECH This research was supported by the National Research Foundation (NRF), funded by the Korean government (MSIT), Medical Device Innovation Center, and the Synthetic Biology Human Resources Development Program. Pohang University of Science & Technology (POSTECH) Journal reference: Park, M., et al. (2025). Drug toxicity prediction based on genotype-phenotype differences between preclinical models and humans. eBioMedicine. doi: 10.1016/j.ebiom.2025.105994. https://www.sciencedirect.com/science/article/pii/S2352396425004384?via%3Dihub
[2]
AI model predicts drug toxicity using differences between animal models and humans
In the UK, there was a case where TGN1412, an immunotherapy under development, triggered a cytokine storm within hours of administration to humans, leading to multiple organ failure. Another example, Aptiganel, a stroke drug candidate, was also highly effective in animals but was discontinued in humans due to side effects such as hallucinations and sedation. Drugs considered safe in preclinical tests can be fatal in human clinical trials. A machine-learning-based technology has been developed to learn these differences and preemptively identify potentially dangerous drugs before clinical trials. A research team led by Professor Sanguk Kim of the Department of Life Sciences and the Graduate School of Artificial Intelligence at POSTECH, along with Dr. Minhyuk Park and Mr. Woomin Song of the Department of Life Sciences, and Mr. Hyunsoo Ahn of the Graduate School of Artificial Intelligence, has developed a technology that uses machine learning to predict drug side effects in humans. The study is published online in eBioMedicine. During the development of new drugs, those that pass preclinical trials often show unexpected toxicity in humans. This issue arises from differences in biological responses between humans and animals. For example, chocolate is generally safe for humans but toxic to dogs. Similarly, a drug that is safe in mice does not necessarily mean it is safe for humans. To date, this "cross-species difference" has been a major reason for failures in new drug development. The research team focused on the "Genotype-Phenotype Difference (GPD)," the biological differences between cells, mice, and humans. They analyzed how genes targeted by drugs function differently in humans and preclinical models, focusing on three key factors: first, the gene's perturbation impact on survival (essentiality); second, the pattern of gene expression in different tissues; and third, the connectivity of genes within biological networks. Validation using data from 434 hazardous drugs and 790 approved drugs revealed that GPD characteristics were significantly associated with drug failure due to toxicity in humans. Predictive power was significantly improved over relying on drug chemical data, with the area under the curve (AUPRC1) increasing from 0.35 to 0.63, and the area under the curve (AUROC2) increasing from 0.50 to 0.75. The developed AI model demonstrated superior predictive performance compared to existing state-of-the-art models. Furthermore, it demonstrated practicality in "chronological validation," which alerts users to drugs facing market withdrawal due to toxicity. After training the prediction model on only drug data up to 1991, it correctly predicted drugs expected to be withdrawn from the market after 1991, achieving 95% accuracy. The significance of this study is that it bridges the "translation gap" between preclinical and clinical trials by quantifying biological differences in cells, preclinical animal models, and humans. Pharmaceutical companies can reduce development costs and time by screening out high-risk candidates before clinical trials, while also improving patient safety. The model's effectiveness is expected to increase as more relevant data and annotations accumulate. Professor Sanguk Kim stated, "This is the first attempt to incorporate differences in genotype-phenotype relationships for drug toxicity prediction. Our framework enables early identification of high-risk drugs in clinical development." He added, "This approach holds promise for reducing development costs, improving patient safety, and increasing the success rate of therapeutic approvals. Co-first authors Dr. Min-hyuk Park and Mr. Woomin Song stated, "The human-centered toxicity prediction model will be a very practical tool in new drug development. We anticipate that pharmaceutical companies will be able to screen out high-risk drugs in advance at the preclinical stage, thereby improving development efficiency."
Share
Share
Copy Link
Researchers at POSTECH have developed a machine learning model that predicts drug toxicity in humans by analyzing biological differences between animal models and humans, potentially preventing dangerous clinical trial failures.
Revolutionary AI Model Addresses Critical Gap in Drug Development
Researchers at Pohang University of Science & Technology (POSTECH) have developed a groundbreaking machine learning model that could fundamentally transform drug safety testing by predicting human toxicity before clinical trials begin
1
. The research, published in eBioMedicine, addresses one of the pharmaceutical industry's most persistent challenges: the translation gap between animal testing and human clinical trials2
.The urgency of this innovation is underscored by historical drug development failures. TGN1412, an immunotherapy candidate, triggered a cytokine storm within hours of human administration, leading to multiple organ failure. Similarly, Aptiganel, a stroke drug that showed promise in animal studies, was discontinued due to severe human side effects including hallucinations and sedation
1
.Understanding Genotype-Phenotype Differences
The research team, led by Professor Sanguk Kim from POSTECH's Department of Life Sciences and Graduate School of Artificial Intelligence, focused on quantifying "Genotype-Phenotype Differences (GPD)" between humans and preclinical models
2
. This approach represents the first attempt to incorporate these biological differences into drug toxicity prediction.The team analyzed three critical factors: gene perturbation impact on survival (essentiality), tissue-specific gene expression patterns, and gene connectivity within biological networks. This comprehensive analysis helps explain why drugs safe in animal models can prove dangerous in humans, similar to how chocolate is safe for humans but toxic to dogs
1
.
Source: Medical Xpress
Impressive Performance Metrics
The AI model demonstrated remarkable predictive capabilities when validated against data from 434 hazardous drugs and 790 approved medications. The system significantly outperformed traditional chemical-based prediction methods, increasing the area under the precision-recall curve (AUPRC) from 0.35 to 0.63 and the area under the receiver operating characteristic curve (AUROC) from 0.50 to 0.75
2
.Perhaps most impressively, the model achieved 95% accuracy in chronological validation tests. When trained only on drug data available through 1991, it successfully predicted which drugs would later be withdrawn from the market due to toxicity concerns
1
.Related Stories
Industry Impact and Future Applications
The practical implications for pharmaceutical companies are substantial. By identifying high-risk drug candidates before expensive clinical trials begin, companies could significantly reduce development costs and timelines while improving patient safety. Co-first authors Dr. Minhyuk Park and Woomin Song emphasized that this "human-centered toxicity prediction model will be a very practical tool in new drug development"
2
.The research was supported by South Korea's National Research Foundation, Medical Device Innovation Center, and Synthetic Biology Human Resources Development Program, indicating strong governmental backing for this innovative approach to drug safety
1
.References
Summarized by
Navi
[1]
Related Stories
Cellarity Unveils AI-Powered ToxPredictor Model to Revolutionize Drug Safety Testing
13 Nov 2025•Science and Research

AI Revolutionizes Drug Discovery: KAIST's BInD Model Designs Optimal Cancer-Fighting Molecules
12 Aug 2025•Science and Research

AI-Driven Drug Discovery Accelerates as FDA Pushes to Reduce Animal Testing
02 Sept 2025•Health

Recent Highlights
1
OpenAI secures $110 billion funding round from Amazon, Nvidia, and SoftBank at $730B valuation
Business and Economy

2
Pentagon accepts OpenAI's autonomous weapons restrictions after blacklisting Anthropic
Policy and Regulation

3
Trump orders federal agencies to ban Anthropic after Pentagon dispute over AI surveillance
Policy and Regulation





