AI Uncovers Promising Antibiotic Compounds in Ancient Archaea Microbes
3 Sources
3 Sources
[1]
Hot springs' hardy microbes offer new source of antibiotics
The mysterious branch of life of known as archaea harbor unique bacteria-fighting compounds Archaea, microbes that make up the least known branch on the tree of life, are home to a trove of leads for new antibiotics, according to a pair of reports out this week. Renowned for their ability to thrive in extreme environments, such as boiling hot springs and salt flats, archaea also exist alongside bacteria in many environments. Now, two groups suggest that proximity may have prompted hundreds of species of archaea to evolve unique chemical defenses, some of which kill bacteria that can sicken people and are resistant to conventional antibiotics. Although the archaeal compounds remain far from proven medicines, the findings underscore that these microbes may wind up becoming a rich vein for antibiotic explorers to mine. The papers, published in Nature Microbiology and PLOS Biology, are "marvelous," says Jim Collins, a bioengineer at the Massachusetts Institute of Technology focused on antibiotic discovery. "I'm surprised we hadn't looked at archaea before." Once thought to be bacteria themselves, archaea are now recognized as a separate domain of life, based on genetic and biochemical differences with bacteria and with eukaryotes, which include fungi and multicellular life. Archaea, for example, do not have the same kind of outer cell wall as bacteria and lack eukaryotes' separate cell nucleus and membrane-bound organelles. Yet, archaea, bacteria, and single-celled eukaryotes all live alongside one another in many environments, such as in the digestive tracts of people, where they help produce methane, and in the hindguts of termites, where they aid in digesting wood. "If they live together in the same niche, chances are they don't always get along," says Tobias Warnecke, a biochemist at the University of Oxford. To see whether potential conflict prodded archaea to develop an antibiotic armamentarium, researchers led by Cesar de la Fuente Nunez, a biomolecular engineer at the University of Pennsylvania, trained an artificial intelligence (AI) algorithm to scan their proteomes -- the complete amino acid sequence of all an organism's proteins. They searched for protein fragments called encrypted peptides that often have antimicrobial properties and are commonly created when larger proteins are broken down. Scanning the proteomes of 233 archaea species revealed more than 12,600 likely encrypted peptides, Nunez and colleagues report this week in Nature Microbiology. The AI also scored each find for its likelihood of having antimicrobial activity, and Nunez's team then synthesized 80 of the most promising peptides. The group found that 93% of these showed antibacterial activity in vitro against dangerous human pathogens such as Staphylococcus aureus and Klebsiella pneumoniae. Further in vitro studies showed most of these peptides kill bacteria by depolarizing their inner, cytoplasmic membranes, a very different strategy than for many known antimicrobial peptides, which more commonly poke holes in bacterial cells' outer membrane. Collins calls the mechanism "a very interesting discovery." Finally, Nunez's team showed in mice that one peptide, called archaeasin-73, reduced loads of dangerous wound-infecting bacteria as much as polymyxin B, a last-line antibiotic treatment. In a separate effort described today in PLOS Biology, Warnecke and his colleagues came at the antimicrobial hunt in archaea from a different direction. Bacteria, unlike archaea, shroud their cell cytoplasmic membranes with a wall formed from a meshlike assembly of peptides called peptidoglycan. So, they wondered whether archaea harbor peptidoglycan-degrading enzymes called hydrolases to defend themselves against bacterial encroachment on their territory. Indeed, the group found these enzymes in 5% of the more than 3700 archaea species they surveyed. Studies of the proteins' structure suggest many are secreted outside the cell, and Warnecke's team even found evidence that some archaea have syringelike molecular injectors for delivering these proteins to bacteria. When exposed to bacteria in the lab, some of the hydrolases shredded their peptidoglycan and killed them. Warnecke and Nunez caution that the newly identified enzymes and peptides remain far from actual drugs that could save the lives of patients. Nevertheless, both argue there are almost certainly many more antibiotics in archaea waiting to be discovered. Warnecke says, "This is probably just the tip of the iceberg."
[2]
AI uncovers 'archaeasins,' unique antibiotics from ancient Archaea
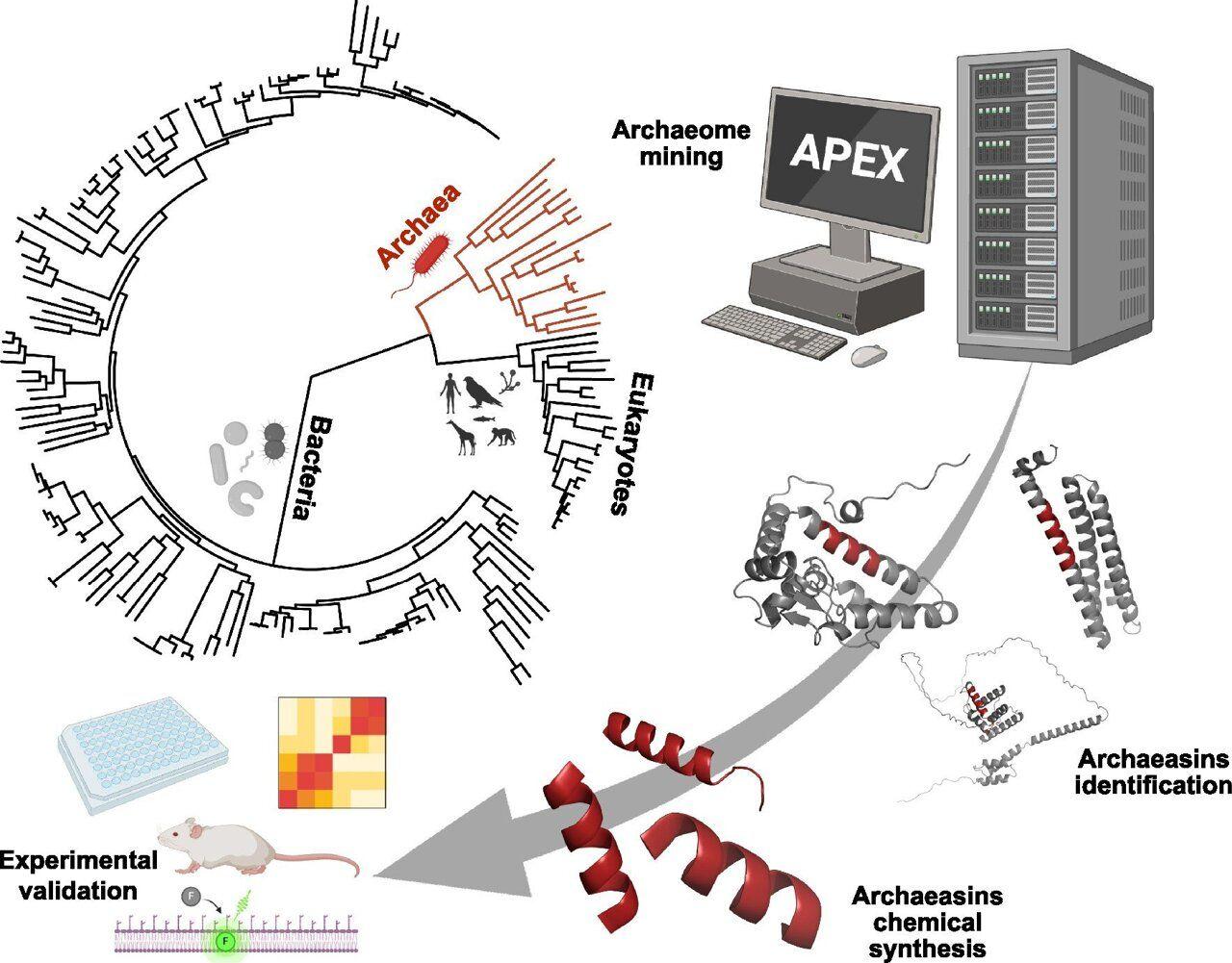
They've survived for billions of years in boiling acid, deep-sea vents and salt flats. Now, some of Earth's oldest life forms -- microbes called Archaea -- are offering a new weapon in the fight against one of today's most urgent health threats: antibiotic resistance. In a study published in Nature Microbiology, researchers at the University of Pennsylvania used artificial intelligence to identify previously unknown compounds in Archaea that could fuel the development of next-generation antibiotics. "Previous efforts to find new antibiotics have looked mostly at fungi, bacteria and animals," says César de la Fuente, Presidential Associate Professor in Bioengineering and in Chemical and Biomolecular Engineering at the University of Pennsylvania School of Engineering and Applied Science (Penn Engineering), in Psychiatry and Microbiology in the Perelman School of Medicine and in Chemistry in the School of Arts & Sciences, and the paper's senior author. In the past, de la Fuente's lab has used AI models to identify antibiotic candidates in a range of unlikely sources, from the DNA of extinct organisms to the chemicals in animal venom. Now, they're applying those tools to a new set of data: the proteins of hundreds of ancient microbes. "There's a whole other domain of life waiting to be explored," says de la Fuente. Exploring a microbial frontier Distinct from both bacteria and from eukaryotes (which include plants, animals and fungi), Archaea occupy their very own branch on the tree of life. Though they resemble bacteria under a microscope, Archaea fundamentally differ in their genetics, cell membranes and biochemistry. These differences allow them to survive in some of Earth's most extreme environments, from superheated undersea vents to blistering hot springs like those in Yellowstone National Park. Because Archaea often thrive where few other organisms can -- enduring crushing pressures, toxic chemicals and extreme temperatures -- their biology has evolved in unusual ways. That makes them a promising but largely untapped source of new molecular tools, including compounds that may function like antibiotics but operate differently from those currently in use. "We were drawn to Archaea because they've had to evolve biochemical defenses in unusual environments," says Marcelo Torres, a research associate in de la Fuente's lab and the paper's co-first author. "We thought, if they've survived for billions of years under those conditions, maybe they've developed unique ways to fight off microbial competitors, and maybe we could learn from that." Hunting for antibiotics with AI To uncover potential antibiotic compounds hidden in Archaea, the researchers turned to artificial intelligence. The team leveraged an updated version of APEX, an AI tool that de la Fuente's lab originally developed to identify antibiotic candidates in ancient biology, including in the proteins of extinct animals like the woolly mammoth. Having seen thousands of peptides -- short chains of amino acids -- with known antimicrobial properties, APEX can predict the likelihood that a given sequence of amino acids will have similar effects. By retraining APEX 1.1 on thousands of additional peptides and information about bacteria that cause diseases in humans, the researchers prepared the tool to predict which peptides in Archaea might inhibit bacterial growth. Scanning 233 species of Archaea yielded more than 12,000 antibiotic candidates. The researchers dubbed these molecules "archaeasins," which chemical analysis revealed differ from known antimicrobial peptides (AMPs), in particular in their distribution of electric charge. The researchers then selected 80 archaeasins to test against actual bacteria. "Trying to find new antibiotics one molecule at a time is like looking for needles in a haystack," says Fangping Wan, a postdoctoral fellow in de la Fuente's lab and the paper's other co-first author. "AI speeds up the process by identifying where the needles are likely to be." On par with existing antibiotics Antibiotics work in a number of ways. Some punch holes in bacterial membranes, while others shut down the organisms' ability to make proteins. The researchers found that, unlike most known AMPs, which attack a bacterium's outer defenses, archaeasins seem to pull the plug from the inside, scrambling the electrical signals that keep the cell alive. In tests against a range of disease-causing, drug-resistant bacteria, 93% of the 80 archaeasins surveyed demonstrated antimicrobial activity against at least one bacterium. The researchers then selected three archaeasins to test in animal models. Four days after a single dose, the archaeasins all arrested the spread of a drug-resistant bacterium often acquired in hospitals. One of the three compounds demonstrated activity comparable to polymyxin B, an antibiotic commonly used as a last-line of defense against drug-resistant infections. "This research shows that there are potentially many antibiotics waiting to be discovered in Archaea," says de La Fuente. "With more and more bacteria developing resistance to existing antibiotics, it's critical to find new antibiotics in unconventional places to replace them." Next, the researchers plan to further enhance APEX so that it can predict antibiotic candidates based on their structure, potentially improving the tool's accuracy. The researchers also hope to better understand the long-term efficacy and safety of archaeasins, with the goal of one day bringing them to human clinical trials. "This is only the beginning," says de la Fuente. "Archaea is one of the oldest forms of life, and clearly has much to teach us about how to outsmart the pathogens we face today."
[3]
AI discovers promising antibiotic compounds in Archaea microbes
University of Pennsylvania School of Engineering and Applied ScienceAug 12 2025 They've survived for billions of years in boiling acid, deep-sea vents and salt flats. Now, some of Earth's oldest life forms - microbes called Archaea - are offering a new weapon in the fight against one of today's most urgent health threats: antibiotic resistance. In a new study published in Nature Microbiology, researchers at the University of Pennsylvania used artificial intelligence to identify previously unknown compounds in Archaea that could fuel the development of next-generation antibiotics. "Previous efforts to find new antibiotics have looked mostly at fungi, bacteria and animals," says César de la Fuente, Presidential Associate Professor in Bioengineering and in Chemical and Biomolecular Engineering in the University of Pennsylvania School of Engineering and Applied Science (Penn Engineering), in Psychiatry and Microbiology in the Perelman School of Medicine and in Chemistry in the School of Arts & Sciences, and the paper's senior author. In the past, de la Fuente's lab has used AI models to identify antibiotic candidates in a range of unlikely sources, from the DNA of extinct organisms to the chemicals in animal venom. Now, they're applying those tools to a new set of data: the proteins of hundreds of ancient microbes. "There's a whole other domain of life waiting to be explored," says de la Fuente. Exploring a microbial frontier Distinct from both bacteria and from eukaryotes (which include plants, animals and fungi), Archaea occupy their very own branch on the tree of life. Though they resemble bacteria under a microscope, Archaea fundamentally differ in their genetics, cell membranes and biochemistry. These differences allow them to survive in some of Earth's most extreme environments, from superheated undersea vents to blistering hot springs like those in Yellowstone National Park. Because Archaea often thrive where few other organisms can - enduring crushing pressures, toxic chemicals and extreme temperatures - their biology has evolved in unusual ways. That makes them a promising but largely untapped source of new molecular tools, including compounds that may function like antibiotics but operate differently from those currently in use. "We were drawn to Archaea because they've had to evolve biochemical defenses in unusual environments," says Marcelo Torres, a research associate in de la Fuente's lab and the paper's co-first author. "We thought, if they've survived for billions of years under those conditions, maybe they've developed unique ways to fight off microbial competitors, and maybe we could learn from that." Hunting for antibiotics with AI To uncover potential antibiotic compounds hidden in Archaea, the researchers turned to artificial intelligence. The team leveraged an updated version of APEX, an AI tool that de la Fuente's lab originally developed to identify antibiotic candidates in ancient biology, including in the proteins of extinct animals like the woolly mammoth. Having seen thousands of peptides - short chains of amino acids - with known antimicrobial properties, APEX can predict the likelihood that a given sequence of amino acids will have similar effects. By retraining APEX 1.1 on thousands of additional peptides and information about bacteria that cause diseases in humans, the researchers prepared the tool to predict which peptides in Archaea might inhibit bacterial growth. Scanning 233 species of Archaea yielded more than 12,000 antibiotic candidates. The researchers dubbed these molecules "archaeasins," which chemical analysis revealed differ from known antimicrobial peptides (AMPs), in particular in their distribution of electric charge. The researchers then selected 80 archaeasins to test against actual bacteria. "Trying to find new antibiotics one molecule at a time is like looking for needles in a haystack," says Fangping Wan, a postdoctoral fellow in de la Fuente's lab and the paper's other co-first author. "AI speeds up the process by identifying where the needles are likely to be." On par with existing antibiotics Antibiotics work in a number of ways. Some punch holes in bacterial membranes, while others shut down the organisms' ability to make proteins. The researchers found that, unlike most known AMPs, which attack a bacterium's outer defenses, archaeasins seem to pull the plug from the inside, scrambling the electrical signals that keep the cell alive. In tests against a range of disease-causing, drug-resistant bacteria, 93% of the 80 archaeasins surveyed demonstrated antimicrobial activity against at least one bacterium. The researchers then selected three archaeasins to test in animal models. Four days after a single dose, the archaeasins all arrested the spread of a drug-resistant bacterium often acquired in hospitals. One of the three compounds demonstrated activity comparable to polymyxin B, an antibiotic commonly used as a last-line of defense against drug-resistant infections. This research shows that there are potentially many antibiotics waiting to be discovered in Archaea. With more and more bacteria developing resistance to existing antibiotics, it's critical to find new antibiotics in unconventional places to replace them." César de la Fuente, Presidential Associate Professor in Bioengineering and in Chemical and Biomolecular Engineering in the University of Pennsylvania School of Engineering and Applied Science Future studies Next, the researchers plan to further enhance APEX so that it can predict antibiotic candidates based on their structure, potentially improving the tool's accuracy. The researchers also hope to better understand the long-term efficacy and safety of archaeasins, with the goal of one day bringing them to human clinical trials. "This is only the beginning," says de la Fuente. "Archaea is one of the oldest forms of life, and clearly has much to teach us about how to outsmart the pathogens we face today." University of Pennsylvania School of Engineering and Applied Science Journal reference: Torres, M. D. T., et al. (2025). Deep learning reveals antibiotics in the archaeal proteome. Nature Microbiology. doi.org/10.1038/s41564-025-02061-0.
Share
Share
Copy Link
Researchers at the University of Pennsylvania have used AI to identify potential new antibiotics from Archaea, ancient microorganisms that thrive in extreme environments. These compounds, dubbed "archaeasins," show promise in fighting drug-resistant bacteria.
AI-Powered Discovery of Novel Antibiotics from Archaea
In a groundbreaking study published in Nature Microbiology, researchers at the University of Pennsylvania have utilized artificial intelligence to uncover a new source of potential antibiotics from one of Earth's oldest life forms - Archaea
1
. This discovery could provide a crucial weapon in the ongoing battle against antibiotic resistance, a pressing global health concern.Exploring the Untapped Potential of Archaea

Source: Science.org
Archaea, distinct from both bacteria and eukaryotes, occupy a unique branch on the tree of life. These microorganisms have evolved to survive in extreme environments, including boiling acid, deep-sea vents, and salt flats
2
. Their ability to thrive in such harsh conditions has led to the development of unique biochemical defenses, making them a promising but largely unexplored source of new molecular tools, including potential antibiotics1
.AI-Driven Antibiotic Discovery
The research team, led by César de la Fuente, employed an updated version of their AI tool called APEX to scan the proteomes of 233 Archaea species
1
3
. This analysis yielded more than 12,000 potential antibiotic candidates, which the researchers named "archaeasins"2
. The AI tool, trained on thousands of peptides with known antimicrobial properties, was able to predict the likelihood of a given amino acid sequence having antibacterial effects3
.Related Stories
Promising Results in Laboratory Tests
Source: Phys.org
Out of the 80 archaeasins selected for testing against actual bacteria, an impressive 93% demonstrated antimicrobial activity against at least one bacterium
2
. Unlike most known antimicrobial peptides that attack a bacterium's outer defenses, archaeasins appear to disrupt the cell from the inside by scrambling electrical signals1
.In animal model tests, three selected archaeasins showed significant efficacy in arresting the spread of a drug-resistant bacterium commonly acquired in hospitals. Notably, one of these compounds demonstrated activity comparable to polymyxin B, an antibiotic often used as a last line of defense against drug-resistant infections
2
.Future Prospects and Challenges

Source: News-Medical
While these findings are promising, the researchers caution that the newly identified compounds are still far from becoming actual drugs that could save patients' lives
1
. The team plans to enhance their AI tool further to predict antibiotic candidates based on structure, potentially improving accuracy3
. They also aim to better understand the long-term efficacy and safety of archaeasins, with the ultimate goal of bringing them to human clinical trials2
.This research not only highlights the potential of AI in drug discovery but also underscores the importance of exploring unconventional sources in the search for new antibiotics. As bacteria continue to develop resistance to existing antibiotics, the exploration of ancient life forms like Archaea may prove crucial in addressing this global health challenge
1
2
3
.References
Summarized by
Navi
[1]
[3]
Related Stories
AI-Designed Antibiotics Show Promise Against Drug-Resistant Superbugs
15 Aug 2025•Science and Research

AI Uncovers Hundreds of Potential Antibiotics in Animal Venom, Offering New Hope Against Antibiotic Resistance
15 Jul 2025•Science and Research

AI Accelerates Antibiotic Discovery: New Drug Targets IBD with Unprecedented Precision
03 Oct 2025•Health

Recent Highlights
1
OpenAI secures $110 billion funding round from Amazon, Nvidia, and SoftBank at $730B valuation
Business and Economy

2
Pentagon accepts OpenAI's autonomous weapons restrictions after blacklisting Anthropic
Policy and Regulation

3
Trump orders federal agencies to ban Anthropic after Pentagon dispute over AI surveillance
Policy and Regulation